Histone O-GlcNAcylation and Potential Biological Functions
Mitsuko Hirosawa 1, *![]() , Koji Hayakawa 1
, Koji Hayakawa 1![]() , Kunio Shiota 2
, Kunio Shiota 2![]() , Satoshi Tanaka 1
, Satoshi Tanaka 1![]()
- Laboratory of Cellular Biochemistry, Department of Animal Resource Sciences/Veterinary Medical Sciences, The University of Tokyo, Tokyo 113-8657, Japan
- Waseda Research Institute for Science and Engineering, Waseda University, Tokyo 169-8555, Japan
* Correspondence: Mitsuko Hirosawa![]()
Received: July 05, 2018 | Accepted: August 30, 2018 | Published: September 19, 2018
OBM Genetics 2018, Volume 2, Issue 3 doi:10.21926/obm.genet.1803036
Academic Editors: Stéphane Viville and Marcel Mannens
Special Issue: Epigenetic Mechanisms in Health and Disease
Recommended citation: Sheth F,Hirosawa M,Hayakawa K,Shiota K, Tanaka S. Histone O-GlcNAcylation and Potential Biological Functions. OBM Genetics 2018;2(3):036; doi:10.21926/obm.genet.1803036.
© 2018 by the authors. This is an open access article distributed under the conditions of the Creative Commons by Attribution License, which permits unrestricted use, distribution, and reproduction in any medium or format, provided the original work is correctly cited.
Abstract
Histone modifications play an important role in the control of DNA-based processes by altering the structure and function of chromatin. O-linked N-acetylglucosamine (O-GlcNAc) modification is a form of post-translational modification of proteins that affects the serine (Ser)/threonine (Thr) residues. This process is controlled by a single pair of enzymes, i.e. O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). Recent evidence indicates the existence of O-GlcNAc modification of histones, with 16 histone O-GlcNAc sites reported to date. O-GlcNAc modification is a nutrient-sensitive modification; therefore, it is likely to serve as a molecular mechanism linking nutrient conditions and epigenetic status. Recently, functional analyses have been advanced by the acquisition of antibodies for the specific detection of O-GlcNAcylation of histone residues. Here, we discuss the current knowledge of histone O-GlcNAc modification, with a view to elucidating its comprehensive biological functions.
Keywords
Histone modification; O-GlcNAcylation; chromatin
1. Introduction
Chromatin, which is a complex consisting mainly of histone proteins and DNA, is indispensable for packaging of the entire genome [1,2,3]. The compaction status of chromatin governs the accessibility of the transcriptional machinery to DNA; thus, it has plays a crucial role in establishing gene expression patterns [4,5]. The nucleosome is the building block of chromatin, containing approximately ~147 base pairs of DNA wrapped around a histone octamer consisting of 2 two copies each of the histones H2A, H2B, H3, and H4 [6,7]. Post-translational modifications (PTMs) of histones may regulate histone-histone and/or histone-DNA interactions related to genome activity, including transcription. Histones are subject to numerous types of PTMs with acetylation, methylation, and phosphorylation being the most abundant modifications forms [8.9]. Recently, accumulating evidence has revealed that histones are also modified by a the addition of a monosaccharide, GlcNAc (Table 1). In this review, we provide an overview of the current understanding of histone O-GlcNAcylation and discuss its potential roles in biological functions.
2. Regulation of O-GlcNAcylation
The nucleotide sugar UDP-GlcNAc serves as a donor for O-GlcNAcylation of Ser/Thr residues of nuclear and cytosolic proteins, which was first reported over 30 years ago [18]. Unlike complex N- and O-linked glyco-chains, the GlcNAc moiety is not further glycocylated following O-GlcNAcylation [19,20]. Compared to other PTMs, the on/off cycling of O-GlcNAcylation of various types of proteins is uniquely regulated by a single pair of enzymes, i.e. O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) [21,22,23,24,25] (Figure 1)
The mechanism by which a single OGT enzyme processes the O-GlcNAcylation of thousands of substrates is believed to be dependent on the protein-protein interaction domain of OGT, the N-terminal tetratricopeptide-repeat (TPR) [23,26]. Although the large diversity of OGT substrates is thought to be achieved by interactions between the TPR of OGT and so-called adaptor proteins, this mechanism is not yet fully understood [22,32,33,34,35,36,37].
In humans and mice, two isoforms of OGA are produced by the alternative splicing of MGEA5/Mgea5 mRNA [27]. The longer isoform, full-length OGA, is composed of an N-terminal GlcNAc hydrolytic cleavage domain for the removal of O-GlcNAc from target proteins [28]. Although there is a C-terminal acetyltransferase-like domain that shares sequence homology with GCN5 histone acetyltransferase (HAT) [29,30], there are discrepancies among several reports on the HAT activity of OGA [38,39]. Moreover, since structural analysis indicates that the C-terminal acetyltransferase-like domain lacks essential residues for the binding of acetyl-CoA as a substrate [40], it is likely that the acetyltransferase-like domain has no HAT activity. However, overexpression studies have confirmed the dual function of Mgea5/OGA as HAT and O-GlcNAcase in neuronal differentiated mouse ES cells [41]. Given the lack of an acetyl-CoA-binding site on OGA itself, it can be speculated that OGA interacts with other proteins with HAT activity. Furthermore, the dual function of Mgea5/OGA is of great interest as an example of crosstalk between histone modifications.
/obm-genetics-0169-20180917-wendy_html_27599ad9.jpg)
Figure 1 O-GlcNAcylation enzymes. The on/off cycling of O-GlcNAc on Ser/Thr residues is catalyzed by a writer (OGT) and an eraser (OGA) enzyme. Alternative splicing produces three isoforms of OGT with different lengths of tetratricopeptide-repeats (TPRs). Nucleocytoplasmic OGT (ncOGT) contains 13 TPRs, whereas mitochondrial OGT (mOGT) and short OGT (sOGT) contain nine and two TPRs, respectively [23,26]. mOGT and sOGT have been implicated in apoptosis. Two isoforms of OGA are produced by alternative splicing [27]. The full-length OGA consists of catalytic and HAT-like domains. The short OGA isoform (sOGA) has a catalytic domain but no HAT-like domain [28,29,30]. Instead, the C-terminal domain of sOGA functions in targeting sOGA to intracellular lipid droplets for regulation of lipid storage [31].
As O-GlcNAcylation occurs on the Ser/Thr residues that are phosphorylated, crosstalk between these two PTMs is suggested to occur through reciprocal competitive block of sites involved in functional switching [42,43,44]. However, when >800 phosphorylation sites were monitored following inhibition of OGA, elevated O-GlcNAcylation resulted in lower phosphorylation at approximately 30% of the monitored sites, and unexpectedly caused increased phosphorylation at approximately 20% of the sites [45]. Therefore, crosstalk between O-GlcNAcylation and phosphorylation cannot be explained only by competitive substitution of both modifications with the same amino acid residue.
3. O-GlcNAcylation is a Nutrient-Sensitive PTM
The interactions between nutrient-sensing mechanisms, such as hexosamine biosynthesis pathway (HBP), and cellular pathways are key to energy homeostasis [46,47]. Less than 5% of total cellular glucose flows into the HBP, leading to the production of UDP-GlcNAc, which is a donor for O-GlcNAcylation [48].
As the HBP depends on the availability of glucose, increasing extracellular glucose elevates the flux through the HBP and results in increased UDP-GlcNAc production. A dynamic change in the O-GlcNAcylated protein pattern is observed even when the concentration of UDP-GlcNAc is only slightly increased due to nutrient excess [49,50,51,52]. Therefore, since cellular levels of UDP-GlcNAc and protein O-GlcNAc fluctuate with the availability of glucose, O-GlcNAcylation is recognized as a nutrient sensor.
As shown in Figure 2, UDP-GlcNAc biosynthesis by the HBP integrates flux not only from carbohydrate metabolism (glucose), but also from other metabolic pathways linked to nutrient intake. Fluctuation in the availability of these nutrients also affects the production of UDP-GlcNAc, causing a dynamic change in GlcNAcylation levels. Thus, O-GlcNAc modification can be considered as a broad range nutrient-sensing PTM [48,53,54].
/obm-genetics-0169-20180917-wendy_html_3ec90315.jpg)
Figure 2 The hexosamine biosynthetic pathway (HBP). The donor for O-GlcNAc modification, UDP-GlcNAc, is the final product of HBP. The HBP integrates metabolites of carbohydrates (glucose), amino acids (glutamine), fatty acids (acetyl-CoA), and nucleotides (UTP) into the synthesis of UDP-GlcNAc, suggesting that the HBP may function as a nutrient-sensing pathway [48,53,54].
4. O-GlcNAcylation is one of a Variety of Histone Modifications
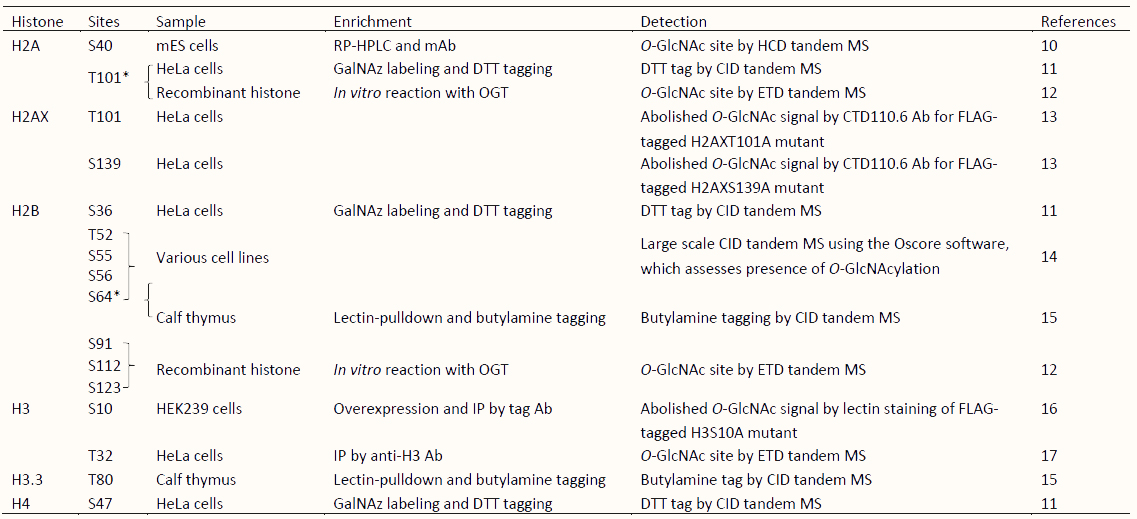
The first study of histone O-GlcNAcylation was reported by Sakabe et al. in 2010 [11]. Since then, 16 histone O-GlcNAcylation sites have been reported (Table 1) [10,12,13,14,15,16,17,55,56,57,58]. Among these, all but two sites were identified by indirect techniques, such as immunoblotting, selective enzymatic labeling, chemoenzymatic detection, and lectin staining, used in combinations with mutation experiments, leaving to skepticism about the true existence of histone O-GlcNAcylation [11,12,13,14,15,16,59]. However, the presence of O-GlcNAc at the two sites, H2AS40 and H3T32, was confirmed based on the detection of endogenous O-GlcNAc by mass spectrometry (MS) analysis of histones isolated from mammalian cells [10,17], even though it is generally acknowledged that direct identification of peptidyl O-GlcNAcylated Ser/Thr by MS is challenging due to its unstable nature. Although the existence of some reported histone O-GlcNAcylations remains controversial [59,60], evidence suggests that O-GlcNAcylation is a form of histone PTM.
Table 1 Overview of published strategies for detection of O-GlcNAc sites on histones.

*O-GlcNAcylated sites identified by independent research. S, Serine; T, Threonine; mES cells, mouse Embryonic Stem cells; RP-HPLC, reversed-phase high performance liquid chromatography; Ab, Antibody; mAb, monoclonal Antibody; GalNAz, azide-modified galactose; DTT, dithiothreitol; IP, immunoprecipitation; MS, mass spectrometry analysis; HCD, higher-energy collisional dissociation; CID, collision-induced dissociation; ETD, electron-transfer dissociation.
5. Biological Functions of O-GlcNAcylated Histones
The histone PTM O-GlcNAcylated H2BS112 was presumed by incubating unmodified histones with OGT and UDP-GlcNAc in vitro. A monoclonal antibody for the specific detection of this modification was originally raised against synthetic O-GlcNAcylated oligopeptides corresponding to a region flanking H2BS112. At present, only polyclonal anti-O-GlcNAcylated H2BS112 antibodies are commercially available. In HeLa cells, H2BS112 O-GlcNAcylation has been shown to promote H2BK120 mono-ubiquitination by providing GlcNAc as an anchor for ubiquitin ligase, leading to transcriptional activation via H3K4me3 [12]. In contrast, O-GlcNAcylation of H2BS112 preserves stable chromatin in the early stages of cell differentiation and may repress gene transcription in adipocytes [56]. Thus, the cellular functions associated with O-GlcNAcylation of H2BS112 appear to be quite diverse.
Mitosis is another cellular event in which histone O-GlcNAcylation is reported to play a role [61,62]. Phosphorylation may occur on the same residues as O-GlcNAcylation; therefore, histone O-GlcNAcylation sites have been functionally analyzed in relation to phosphorylation. H3S10 and H3T32 are known to be phosphorylated when cells enter mitosis [61,62]. The increase in O-GlcNAcylation of H3T32 causes a decrease in mitosis-specific phosphorylation of S10, S28, and T32 [16,17,61,62], indicating that O-GlcNAcylation of H3T32 regulates mitosis by modulating mitosis-related histone phosphorylation. In addition, mitosis-specific phosphorylation of H3S10 can be competitively reduced by the level of O-GlcNAcylation at this site. Therefore, it can be speculated that O-GlcNAcylation of H3S10 regulates the pathways involving H3S10 phosphorylation, such as the G2/M checkpoint [9,16,17].
Functional analysis of O-GlcNAcylation at H2AT101 (H2AT101Gc) has been advanced by nucleosome reconstitution methods using synthesized GlcNAcylated histones. H2AT101Gc has been shown to influence nucleosome structure through destabilization of H2A/H2B dimers, causing the promotion of relaxed chromatin [55].
6. Newly Discovered O-GlcNAc Modification of H2AS40 in Placental Mammals
As part of the search for novel histone O-GlcNAcylation sites, a series of monoclonal antibodies were generated using an O-GlcNAcylated oligopeptide library containing several putative O-GlcNAcylation sites designed based on preliminary MS analyses of purified histones derived from mouse ES cells. One of the obtained antibodies, 20B2, was found to specifically recognize O-GlcNAcylated H2AS40 (H2AS40Gc) [10], which functions to maintain genome integrity through the DNA repair mechanism in coordination with γH2AX and AcH2AZ [58]. As O-GlcNAcylation of H2AXS139 [13] and H2BS112 [57] has also been implicated in DNA repair, we propose a DNA repair response mechanism centered on histone O-GlcNAc modifications.
The majority of histone modifications are highly conserved within the animal kingdom [63]. In contrast, a genomic database survey revealed that H2AS40Gc, with Ser at position 40, instead of Ala in the “universal” H2A, appears to be a modification that is unique to placental mammals. Chromatin immunoprecipitation sequencing (ChIP-seq) for genome-wide localization of H2AS40Gc in mouse trophoblast stem cells showed a dynamic change in the distribution pattern with differentiation [10]. Oxidative stress occurs in a normal placenta with the establishment of maternal circulation [64,65,66]; therefore, placental mammal-specific H2AS40Gc might play a role in genome protection against DNA damage by reactive oxygen species (ROS) produced under oxidative stress in the placental environment. However, further investigations are required to verify this hypothesis.
7. Perspectives on O-GlcNAcylated Histones
As mentioned previously, O-GlcNAcylation-specific antibodies are a very powerful tool for functional analyses [10,12,56,58]. However, historically, it has been difficult to obtain specific antibodies against sugar moieties. Even when such antibodies are successfully produced, they are usually IgM antibodies, which have limited use [67]. IgG antibodies specifically directed against O-GlcNAc histone modifications that are suitable for use in ChIP-seq analysis are required to progress our understanding of O-GlcNAc modification. We have attempted to produce such antibodies; the characterization of these antibodies and the validation of their targets is currently underway.
In hyperglycemia (insulin resistance), a rich UDP-GlcNAc pool produced by HBP flux results in an abnormally high level of protein O-GlcNAcylation. In the case of some key transcription factors or coactivators, increased levels of O-GlcNAcylation are known to stimulate gluconeogenesis/lipogenesis transcription, which further diminishes insulin sensitivity [20,68,69]. Histone O-GlcNAcylation could be regarded as a new factor in insulin resistance through epigenetic regulation. Therefore, investigations of the responses of histone O-GlcNAc modification to hyper-or hypoglycemic status are of great interest, suggensting that O-GlcNAcylated histone might have the potential in the diagnosis and/or prevention of chronic metabolic diseases such as diabetes.
Recent studies have shown that unbalanced O-GlcNAc modification leads not only to metabolic disease [46,70,71], but also to various types of other conditions, including neurological disorders [72,73], cardiovascular disease [74,75], and cancer [76,77,78]. In particular, high levels of O-GlcNAc modifications have been observed in breast [79,80,81], prostate [82,83], lung, and colon cancers [84,85], as well as in hepatocellular carcinoma [86,87]. Although O-GlcNAc modification levels have been typically analyzed using only a pan-O-GlcNAcylation antibody, however, we speculate that histones are the key targets of O-GlcNAcylation and that as epigenetic mechanisms underlie the onset of chronic diseases [88,89,90,91].
The combination of the wide variety of histone modifications allows for increased complexity of epigenetic regulation [8]. With advances such as ChIP-seq, it is now possible to map the genome-wide distribution or colocalization of histone modifications at high resolutions, revealing the many combinations of histone modification-crosstalk, such as mutual exclusion, precondition, or coexistence [92]. The identification of the colocalization of H2BS112Gc and H2BK120 mono-ubiquitination [12], and that of H2S40Gc and γH2AX [58], has contributed to the elucidation of the biological functions of these histone O-GlcNAcylations. Thus, it is worth focusing on the crosstalk between O-GlcNAcylation and other histone modifications with the aim of determining the function of histone O-GlcNAc modifications.
The interplay between phosphorylation and O-GlcNAcylation may be one example of such crosstalk. Competition between phosphorylation and O-GlcNAcylation has been reported only for the H3S10, H3T32, and H2AXS139 sites [13,16,17]. Future studies should, therefore, investigate phosphorylation of other residues for which O-GlcNAc modification has been reported. In addition, considering the presence of a HAT-like domain in OGA, the crosstalk between O-GlcNAcylation and acetylation should be validated, despite the controversy regarding the true HAT enzymatic activity of OGA [18,30,39,40,41,93].
Studies of histone PTMs have focused mainly on the flexible N/C-terminal tails as the targets of numerous functional modifications that act by recruiting effector proteins [94,95]. Whereas modifications in the histone tails might have a limited structural impact on the nucleosome itself, PTMs in the globular domain of histones have a direct structural effect on the nucleosome through their influence on histone-histone or histone-DNA interactions [96,97]. Of the 16 O-GlcNAcylation sites of histones reported, 13 sites are not present in the tail, but are located on the surface of the histone octamer (5 sites) or even on the inside of the nucleosome (8 sites) [10,12,13,14,15,16,17,55,56,57,58,98] (Figure 3). Therefore, it can be hypothesized that most O-GlcNAc modifications of histones function at the level of chromatin dynamics based on the structural changes of the nucleosome. This raises the new question as to whether such internal Ser/Thr residues are O-GlcNAcylated before or after nucleosome assembly. It is possible that O-GlcNAcylation site-specific adapter proteins facilitate the access of OGT to the inside of nucleosomes. Reconstitution experiments using a synthetic pure O-GlcNAcylated histone performed by Lercher et al. for H2AT101 [55], could help to clarify this issue.
/obm-genetics-0169-20180917-wendy_html_e9c25812.jpg)
Figure 3 O-GlcNAcylated histone residues mapped onto the nucleosome structure. Top (left) and lateral (right) view of nucleosome structure (PDB ID: 3AFA) [98]. H2A: pink, H2B: orange, H3: green, H4: blue. The O-GlcNAcylated Ser or Thr side chains are indicated by spheres. O-GlcNAcylated residues in the histone tail: blue spheres, O-GlcNAcylated residues at the nucleosome surface: red spheres, O-GlcNAcylated residues inside the nucleosome: black spheres, reported characteristics.
8. Conclusions
Although the biological functions of histone O-GlcNAcylation have been gradually unveiled since its discovery nearly a decade ago, our knowledge of this protein modification is still limited. On the basis that O-GlcNAcylation functions as a nutrient sensor, histone O-GlcNAcylation can be regarded as a molecular mechanism linking metabolism and epigenetics, thus establishing a new paradigm of the epigenetic basis of chronic metabolic diseases.
Acknowledgments
This work was supported by the Lotte Shigemitsu Prize. We would like to thank Editage (www.editage.jp) for their English language editing.
Author Contributions
M.H. wrote the manuscript with input from all authors.
Competing Interests
The authors declare no competing interests.
References
- Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011; 21: 175-186. [CrossRef] [Google scholar] [PubMed]
- Maeshima K, Imai R, Tamura S, Nozaki T. Chromatin as dynamic 10-nm fibers. Chromosoma. 2014; 123: 225-237. [CrossRef] [Google scholar] [PubMed]
- Ozer G, Luque A, Schlick T. The chromatin fiber: multiscale problems and approaches. Curr Opin Struct Biol. 2015; 31: 124-139. [CrossRef] [Google scholar] [PubMed]
- Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014; 83: 671-696. [CrossRef] [Google scholar] [PubMed]
- Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014; 15: 69-81. [CrossRef] [Google scholar] [PubMed]
- Cutter AR, Hayes JJ. A brief review of nucleosome structure. FEBS Lett. 2017; 589: 2914-2922. [CrossRef] [Google scholar] [PubMed]
- McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015; 115: 2255-2273. [CrossRef] [Google scholar] [PubMed]
- Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014; 15: 703-708. [CrossRef] [Google scholar] [PubMed]
- Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015; 16: 1467-1481. [CrossRef] [Google scholar] [PubMed]
- Hirosawa M, Hayakawa K, Yoneda C, Arai D, Shiota H, Suzuki T, et al. Novel O-GlcNAcylation on Ser(40) of canonical H2A isoforms specific to viviparity. Sci Rep. 2016; 6: 31785. [CrossRef] [Google scholar] [PubMed]
- Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010; 107: 19915-19920. [CrossRef] [Google scholar] [PubMed]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011; 480: 557-560. [CrossRef] [Google scholar] [PubMed]
- Chen Q, Yu X. OGT restrains the expansion of DNA damage signaling. Nucleic Acids Res. 2016; 44: 9266-9278. [CrossRef] [Google scholar] [PubMed]
- Hahne H, Moghaddas Gholami A, Kuster B. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol Cell Proteomics. 2012; 11: 843-850. [CrossRef] [Google scholar] [PubMed]
- Schouppe D, Ghesquiere B, Menschaert G, De Vos WH, Bourque S, Trooskens G, et al. Interaction of the tobacco lectin with histone proteins. Plant Physiol. 2011; 155: 1091-1102. [CrossRef] [Google scholar] [PubMed]
- Zhang S, Roche K, Nasheuer HP, Lowndes NF. Modification of histones by sugar beta-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011; 286: 37483-37495. [CrossRef] [Google scholar] [PubMed]
- Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, et al. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012; 287: 12195-12203. [CrossRef] [Google scholar] [PubMed]
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984; 259: 3308-3317. [Google scholar]
- Zachara N, Akimoto Y, Hart GW. The O-GlcNAc Modification. In: 3rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, et al, editors. Essentials of Glycobiology Cold Spring Harbor (NY): by The Consortium of Glycobiology Editors, La Jolla, California. 2015; p. 239-251. [Google scholar]
- Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015; 208: 869-880. [CrossRef] [Google scholar] [PubMed]
- Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997; 272: 9308-9315. [CrossRef] [Google scholar] [PubMed]
- Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011; 469: 564-567. [CrossRef] [Google scholar] [PubMed]
- Levine ZG, Walker S. The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells? Annu Rev Biochem. 2016; 85: 631-657. [CrossRef] [Google scholar] [PubMed]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001; 276: 9838-9845. [CrossRef] [Google scholar] [PubMed]
- Alonso J, Schimpl M, van Aalten DM. O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J Biol Chem. 2014; 289: 34433-34439. [CrossRef] [Google scholar] [PubMed]
- Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, et al. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003; 409: 287-297. [CrossRef] [Google scholar] [PubMed]
- Comtesse N, Maldener E, Meese E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem Biophys Res Commun. 2001; 283: 634-640. [CrossRef] [Google scholar] [PubMed]
- Haltiwanger RS, Holt GD, Hart GW. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. J Biol Chem. 1990; 265: 2563-2568. [Google scholar]
- Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem. 2004; 279: 53665-53673. [CrossRef] [Google scholar] [PubMed]
- Toleman CA, Paterson AJ, Kudlow JE. The histone acetyltransferase NCOAT contains a zinc finger-like motif involved in substrate recognition. J Biol Chem. 2006; 281: 3918-3925. [CrossRef] [Google scholar] [PubMed]
- Keembiyehetty CN, Krzeslak A, Love DC, Hanover JA. A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J Cell Sci. 2011; 124: 2851-2860. [CrossRef] [Google scholar] [PubMed]
- Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J Biol Chem. 2003; 278: 5399-5409. [CrossRef] [Google scholar] [PubMed]
- Marz P, Stetefeld J, Bendfeldt K, Nitsch C, Reinstein J, Shoeman RL, et al. Ataxin-10 interacts with O-linked beta-N-acetylglucosamine transferase in the brain. J Biol Chem. 2006; 281: 20263-20270. [CrossRef] [Google scholar] [PubMed]
- Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008; 283: 33935-33941. [CrossRef] [Google scholar] [PubMed]
- Deng RP, He X, Guo SJ, Liu WF, Tao Y, Tao SC. Global identification of O-GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. Proteomics. 2014; 14: 1020-1030. [CrossRef] [Google scholar] [PubMed]
- Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, et al. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat Struct Mol Biol. 2015; 22: 744-750. [CrossRef] [Google scholar] [PubMed]
- Rafie K, Raimi O, Ferenbach AT, Borodkin VS, Kapuria V, van Aalten DMF. Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017; 7: 10.1098/rsob.170078. [CrossRef] [Google scholar] [PubMed]
- Kim EJ. In Vitro Biochemical Assays for O-GlcNAc-Processing Enzymes. Chembiochem. 2017; 18: 1462-1472. [CrossRef] [Google scholar] [PubMed]
- Butkinaree C, Cheung WD, Park S, Park K, Barber M, Hart GW. Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J Biol Chem. 2008; 283: 23557-23566. [CrossRef] [Google scholar] [PubMed]
- Rao FV, Schuttelkopf AW, Dorfmueller HC, Ferenbach AT, Navratilova I, van Aalten DM. Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol. 2013; 3: 130021. [CrossRef] [Google scholar] [PubMed]
- Hayakawa K, Hirosawa M, Tabei Y, Arai D, Tanaka S, Murakami N, et al. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J Biol Chem. 2013; 288: 17099-17110. [CrossRef] [Google scholar] [PubMed]
- Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010; 584: 2526-2538. [CrossRef] [Google scholar] [PubMed]
- Hart GW. Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol (Lausanne). 2014; 5: 183. [CrossRef] [Google scholar] [PubMed]
- van der Laarse SAM, Leney AC, Heck AJR. Crosstalk between phosphorylation and O-GlcNAcylation: friend or foe. FEBS J. 2018. doi.org/10.1111/febs.14491 [CrossRef] [Google scholar] [PubMed]
- Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci USA. 2008; 105: 13793-13798. [CrossRef] [Google scholar] [PubMed]
- Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013; 33: 205-229. [CrossRef] [Google scholar] [PubMed]
- Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014; 20: 208-213. [CrossRef] [Google scholar] [PubMed]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991; 266: 4706-4712. [Google scholar]
- Liu J, Marchase RB, Chatham JC. Glutamine-induced protection of isolated rat heart from ischemia/reperfusion injury is mediated via the hexosamine biosynthesis pathway and increased protein O-GlcNAc levels. J Mol Cell Cardiol. 2007; 42: 177-185. [CrossRef] [Google scholar] [PubMed]
- Matthews JA, Belof JL, Acevedo-Duncan M, Potter RL. Glucosamine-induced increase in Akt phosphorylation corresponds to increased endoplasmic reticulum stress in astroglial cells. Mol Cell Biochem. 2007; 298: 109-123. [CrossRef] [Google scholar] [PubMed]
- Jones DR, Keune WJ, Anderson KE, Stephens LR, Hawkins PT, Divecha N. The hexosamine biosynthesis pathway and O-GlcNAcylation maintain insulin-stimulated PI3K-PKB phosphorylation and tumour cell growth after short-term glucose deprivation. FEBS J. 2014; 281: 3591-3608. [CrossRef] [Google scholar] [PubMed]
- Dassanayaka S, Readnower RD, Salabei JK, Long BW, Aird AL, Zheng YT, et al. High glucose induces mitochondrial dysfunction independently of protein O-GlcNAcylation. Biochem J. 2015; 467: 115-126. [CrossRef] [Google scholar] [PubMed]
- Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010; 1800: 96-106. [CrossRef] [Google scholar] [PubMed]
- Love DC, Krause MW, Hanover JA. O-GlcNAc cycling: emerging roles in development and epigenetics. Semin Cell Dev Biol. 2010; 21: 646-654. [CrossRef] [Google scholar] [PubMed]
- Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, et al. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat Commun. 2015; 6: 7978. [CrossRef] [Google scholar] [PubMed]
- Ronningen T, Shah A, Oldenburg AR, Vekterud K, Delbarre E, Moskaug JO, et al. Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 2015; 25: 1825-1835. [CrossRef] [Google scholar] [PubMed]
- Wang P, Peng C, Liu X, Liu H, Chen Y, Zheng L, et al. OGT mediated histone H2B S112 GlcNAcylation regulates DNA damage response. J Genet Genomics. 2015; 42: 467-475. [CrossRef] [Google scholar] [PubMed]
- Hayakawa K, Hirosawa M, Tani R, Yoneda C, Tanaka S, Shiota K. H2A O-GlcNAcylation at serine 40 functions genomic protection in association with acetylated H2AZ or gammaH2AX. Epigenetics Chromatin. 2017; 10: 51. [CrossRef] [Google scholar] [PubMed]
- Gambetta MC, Muller J. A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma. 2015; 124: 429-442. [CrossRef] [Google scholar] [PubMed]
- Gagnon J, Daou S, Zamorano N, Iannantuono NV, Hammond-Martel I, Mashtalir N, et al. Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics. 2015; 10: 677-691. [CrossRef] [Google scholar] [PubMed]
- Caperta AD, Rosa M, Delgado M, Karimi R, Demidov D, Viegas W, et al. Distribution patterns of phosphorylated Thr 3 and Thr 32 of histone H3 in plant mitosis and meiosis. Cytogenet Genome Res. 2008; 122: 73-79. [CrossRef] [Google scholar] [PubMed]
- Perez-Cadahia B, Drobic B, Davie JR. H3 phosphorylation: dual role in mitosis and interphase. Biochem Cell Biol. 2009; 87: 695-709. [Google scholar]
- Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci. 2006; 11: 199-208. [CrossRef] [Google scholar] [PubMed]
- Rodesch F, Simon P, Donner C, Jauniaux E. Oxygen measurements in endometrial and trophoblastic tissues during early pregnancy. Obstet Gynecol. 1992; 80: 283-285. [Google scholar]
- Genbacev O, Joslin R, Damsky CH, Polliotti BM, Fisher SJ. Hypoxia alters early gestation human cytotrophoblast differentiation/invasion in vitro and models the placental defects that occur in preeclampsia. J Clin Invest. 1996; 97: 540-550. [CrossRef] [Google scholar] [PubMed]
- Patel J, Landers K, Mortimer RH, Richard K. Regulation of hypoxia inducible factors (HIF) in hypoxia and normoxia during placental development. Placenta. 2010; 31: 951-957. [CrossRef] [Google scholar] [PubMed]
- Heimburg-Molinaro J, Rittenhouse-Olson K. Development and characterization of antibodies to carbohydrate antigens. Methods Mol Biol. 2009; 534: 341-357. [Google scholar]
- Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013; 10: 365-380. [CrossRef] [Google scholar] [PubMed]
- Vaidyanathan K, Wells L. Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type II diabetes. J Biol Chem. 2014; 289: 34466-34471. [CrossRef] [Google scholar] [PubMed]
- Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab. 2013; 24: 301-309. [CrossRef] [Google scholar] [PubMed]
- Olivier-Van Stichelen S, Hanover JA. You are what you eat: O-linked N-acetylglucosamine in disease, development and epigenetics. Curr Opin Clin Nutr Metab Care. 2015; 18: 339-345. [CrossRef] [Google scholar] [PubMed]
- Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, et al. O-GlcNAc modification blocks the aggregation and toxicity of the protein alpha-synuclein associated with Parkinson's disease. Nat Chem. 2015; 7: 913-920. [CrossRef] [Google scholar] [PubMed]
- Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. The emerging link between O-GlcNAc and Alzheimer disease. J Biol Chem. 2014; 289: 34472-34481. [CrossRef] [Google scholar] [PubMed]
- Dassanayaka S, Jones SP. O-GlcNAc and the cardiovascular system. Pharmacol Ther. 2014; 142: 62-71. [CrossRef] [Google scholar] [PubMed]
- Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, et al. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013; 502: 372-376. [CrossRef] [Google scholar] [PubMed]
- Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011; 11: 678-684. [CrossRef] [Google scholar] [PubMed]
- Ma Z, Vosseller K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J Biol Chem. 2014; 289: 34457-34465. [CrossRef] [Google scholar] [PubMed]
- Singh JP, Zhang K, Wu J, Yang X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015; 356: 244-250. [CrossRef] [Google scholar] [PubMed]
- Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010; 29: 2831-2842. [CrossRef] [Google scholar] [PubMed]
- Krzeslak A, Forma E, Bernaciak M, Romanowicz H, Brys M. Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin Exp Med. 2012; 12: 61-65. [CrossRef] [Google scholar] [PubMed]
- Chaiyawat P, Netsirisawan P, Svasti J, Champattanachai V. Aberrant O-GlcNAcylated Proteins: New Perspectives in Breast and Colorectal Cancer. Front Endocrinol (Lausanne) 2014;5:193. [CrossRef] [Google scholar] [PubMed]
- Gu Y, Gao J, Han C, Zhang X, Liu H, Ma L, et al. O-GlcNAcylation is increased in prostate cancer tissues and enhances malignancy of prostate cancer cells. Mol Med Rep. 2014; 10: 897-904. [CrossRef] [Google scholar] [PubMed]
- Kamigaito T, Okaneya T, Kawakubo M, Shimojo H, Nishizawa O, Nakayama J. Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis. 2014; 17: 18-22. [CrossRef] [Google scholar] [PubMed]
- Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, et al. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011; 1812: 514-519. [CrossRef] [Google scholar] [PubMed]
- Yang YR, Kim DH, Seo YK, Park D, Jang HJ, Choi SY, et al. Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-kappaB signaling. Oncotarget. 2015; 6: 12529-12542. [Google scholar]
- Zhu G, Tao T, Zhang D, Liu X, Qiu H, Han L, et al. O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression. Glycobiology. 2016; 26: 820-833. [CrossRef] [Google scholar] [PubMed]
- Liu Q, Tao T, Liu F, Ni R, Lu C, Shen A. Hyper-O-GlcNAcylation of YB-1 affects Ser102 phosphorylation and promotes cell proliferation in hepatocellular carcinoma. Exp Cell Res. 2016; 349: 230-238. [CrossRef] [Google scholar] [PubMed]
- Slomko H, Heo HJ, Einstein FH. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012; 153: 1025-1030. [CrossRef] [Google scholar] [PubMed]
- Brunet A, Berger SL. Epigenetics of aging and aging-related disease. J Gerontol A Biol Sci Med Sci. 2014; 69: S17-S20. [CrossRef] [Google scholar] [PubMed]
- Reddy MA, Natarajan R. Recent developments in epigenetics of acute and chronic kidney diseases. Kidney Int. 2015; 88: 250-261. [CrossRef] [Google scholar] [PubMed]
- Mann DA. Epigenetics in liver disease. Hepatology. 2014; 60: 1418-1425. [CrossRef] [Google scholar] [PubMed]
- Pick H, Kilic S, Fierz B. Engineering chromatin states: chemical and synthetic biology approaches to investigate histone modification function. Biochim Biophys Acta. 2014; 1839: 644-656. [CrossRef] [Google scholar] [PubMed]
- Kim EJ, Amorelli B, Abdo M, Thomas CJ, Love DC, Knapp S, et al. Distinctive inhibition of O-GlcNAcase isoforms by an alpha-GlcNAc thiolsulfonate. J Am Chem Soc. 2007; 129: 14854-14855. [CrossRef] [Google scholar] [PubMed]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293: 1074-1080. [CrossRef] [Google scholar] [PubMed]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012; 19: 1218-1227. [CrossRef] [Google scholar] [PubMed]
- Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004; 11: 1037-1043. [CrossRef] [Google scholar] [PubMed]
- Tropberger P, Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol. 2013; 20: 657-661. [CrossRef] [Google scholar] [PubMed]
- Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, et al. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc Natl Acad Sci USA. 2010; 107: 10454-10459. [CrossRef] [Google scholar] [PubMed]