Non-Coding RNAs in Cutaneous Melanoma Development, Progression and Dissemination
Virginie Vignard 1,2 ![]() , Delphine Fradin 1,*
, Delphine Fradin 1,*![]()
- CRCINA, Inserm, Université de Nantes, IRSUN 8 quai Moncousu, Nantes, France
- CHU Nantes, Nantes, France
* Correspondence: Delphine Fradin![]()
Received: March 13, 2018 | Accepted: May 06, 2018 | Published: May 16, 2018
OBM Genetics 2018, Volume 2, Issue 2 doi:10.21926/obm.genet.1802020
Academic Editors: Stéphane Viville and Marcel Mannens
Special Issue: Epigenetic Mechanisms in Health and Disease
Recommended citation: Vignard V, Fradin D. Non-Coding RNAs in Cutaneous Melanoma Development, Progression and Dissemination. OBM Genetics 2018;2(2):020; doi:10.21926/obm.genet.1802020.
© 2018 by the authors. This is an open access article distributed under the conditions of the Creative Commons by Attribution License, which permits unrestricted use, distribution, and reproduction in any medium or format, provided the original work is correctly cited.
Abstract
Melanoma is a highly aggressive skin cancer with high incidence worldwide. There is growing evidence that aberrantly expressed non-coding RNAs (ncRNAs) play a role in the development, progression and dissemination of melanoma tumor cells. Among the many types of ncRNAs described in this review, the functions of micro- and long non-coding RNAs are described and related to the six hallmarks of cancer. Recently, ncRNAs discovered in body fluids have become known as one of the most promising groups of oncological biomarkers for use in non-invasive diagnosis, prognosis or therapeutic response prediction.
Keywords
Non-coding RNA; melanoma; cancer hallmarks
1. Introduction
Melanoma is the deadliest form of skin cancer. The prognosis remains favorable for those diagnosed with early stage disease, where surgical therapy is the mainstay of treatment. However, treatment rarely succeeds if begun in the advanced stages of disease, although survival of late-stage disease has considerably improved in recent years [1,2].
Interest in the identification of molecular pathways that contribute to melanoma development and progression has grown significantly over the last few years, revealing new insights for development of targeted melanoma therapies such as BRAF (B-Raf proto-oncogene, serine/threonine kinase) kinase inhibitor or MEK (mitogen-activated protein kinase kinase 1/2) inhibitor [3,4]. Concurrently, the manipulation of the immune system [5] (e.g., through the blocking of immune checkpoints or through adoptive cell transfer) has become an important strategy for controlling disease progression during advanced melanoma stages. Although these therapies show promising clinical results, primary or acquired resistance to therapy is common and clinical outcomes for metastatic disease still remain poor.
Recently, epigenetic modifications have also been implicated in the pathogenesis of many human diseases, including melanoma. The most widely accepted definition of epigenetics includes stably inherited modulations in the expression of genes that do not involve changes in their DNA sequences. In this view, gene expression can vary due to effects exerted by RNA molecules. Specifically, increasing emphasis is being placed on the roles of non-coding RNAs (ncRNAs) to modulate gene expression through their functions as epigenetic modifiers [6]. Since ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) were discovered in the 1950s [7,8], numerous other classes of RNAs located within the nucleus or cytoplasm have joined their ranks, including ncRNAs [9]. The two most described classes of ncRNAs are micro-RNAs (miRNAs) and the very large group of long ncRNAs (lncRNAs). Micro-RNAs, ncRNAs that are 19-22 nucleotides in length, modulate gene expression through mRNA silencing or degradation [10]. Notably, a single miRNA has the capacity to inhibit several different mRNA targets; this feature explains how miRNAs are such powerful regulators of gene expression. Moreover, they are variably expressed by cells and control many pathways that participate in cell metabolism, proliferation, differentiation and apoptosis [11]. Indeed, various studies have demonstrated that more than 60% of human genes are regulated by miRNAs [12] that may act as tumor suppressors (TS-miR) or oncogenes (oncomiR) depending on tissue, cellular context and target genes [13,14]. The non-coding genome is also transcribed into many thousands of lncRNAs that are >200 nucleotides in length [15]. In contrast to miRNAs, which mainly negatively regulate gene expression, lncRNAs may either induce or repress gene expression and are more specific in terms of species, tissue and tumor type.
The objective of this review is to present information with regard to selected micro-RNAs and long non-coding RNAs in melanoma and relate their functions to the six hallmarks of cancer proposed by Hanahan and Weinberg in 2000 [16]. In that work, they suggested that cancer cells are characterized by six essential physiological alterations that are conducive to malignant growth: i) sustaining proliferative signaling; ii) evading growth suppressors; iii) enabling replicative immortality; iv) activating invasion and metastasis; v) inducing angiogenesis and vi) resisting cell death. This is the first review that relates short and long non-coding RNAs to current knowledge with regard to the six cancer hallmarks of cutaneous melanoma.
2. Non-Coding RNAs Maintain Proliferative Signaling in Melanoma Cells
To maintain a proliferative state, cancer cells often produce their own growth factors (GF) while concomitantly overexpressing GF receptors. Subsequently, GF bind to over-expressed GF receptors to induce autocrine proliferative stimulation, thus reducing their dependency on the external microenvironment for stimulation. Melanoma cells can also constitutively activate their own receptors via signals that are transduced within the tumor cells by activation of MAPK (mitogen-activated protein kinase) or NOTCH pathways (Figure 1). Such growth-signaling pathways in cancer cells have been linked to deregulation of the normal activities of non-coding RNA [17].
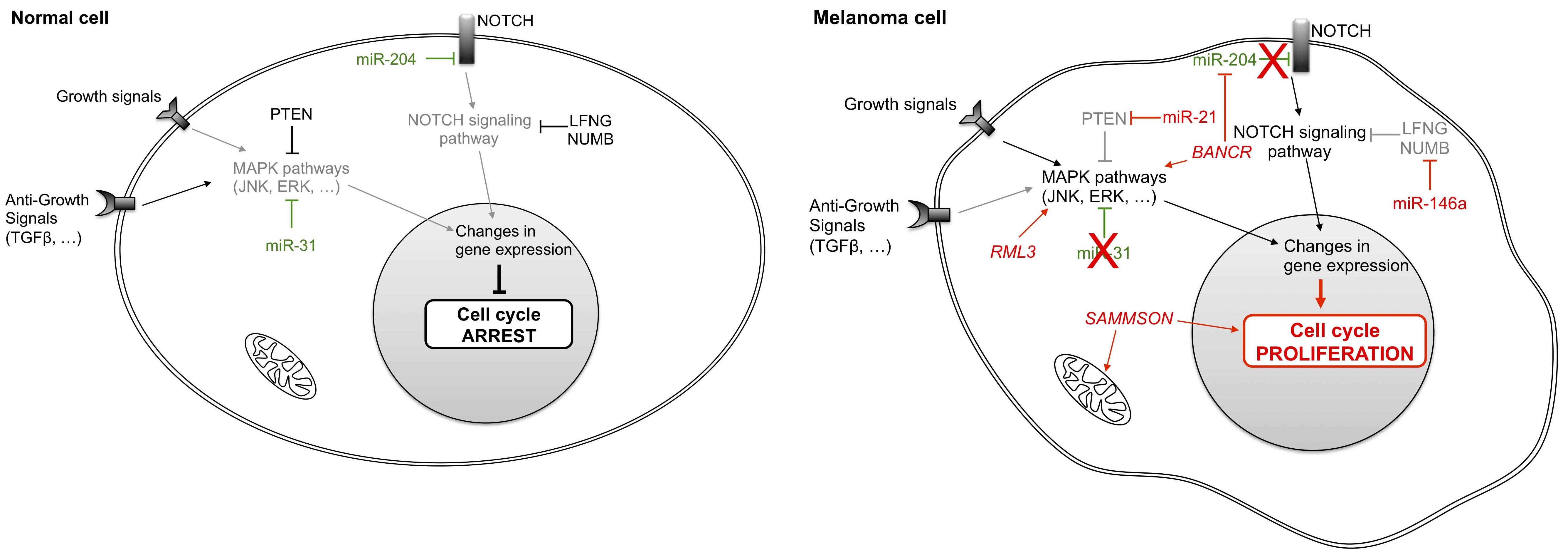
Figure 1 Differential non-coding RNA expression in normal and melanoma cells influences growth signals. Normal cells regulate their growth by modulating the activity of MAPK pathways and of the NOTCH signaling pathway. In tumor cells, aberrant expression of several micro-RNAs and long non-coding RNAs cause deregulation of these pathways and of cell proliferation. LFNG: LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase; NUMB: numb homolog; PTEN: phosphatase and tensin homolog; JNK: mitogen-activated protein kinase 8; ERK: elk-related tyrosine kinase; TGFβ: transforming growth factor beta; BANCR: BRAF-activated non-protein coding RNA; SAMMSON: survival associated mitochondrial melanoma-specific oncogenic non-coding RNA.
Non-coding RNAs can be categorized into two groups. The first group encompasses ncRNAs that normally function as tumor suppressors, but which are down-regulated and therefore not effectively present in cancer cells. For example, in non-tumor cells miR-31 targets kinases such as MET (proto-oncogene receptor tyrosine kinase), RAB27 (Ras-related protein in brain) and NIK (NFKB inducing kinase) to block signaling and inhibit cell proliferation [18]. Expression of each of these kinases is often enhanced in melanoma cells, due to the frequent deletion of the miR-31 gene [19]. Another ncRNA, miR-204, also plays a central role in maintaining a non-proliferative state in noncancerous cells by down-regulating the NOTCH signaling pathway.
Conversely, other non-coding RNAs that act as oncogenes are over-expressed in melanoma cells. The miRNA miR-21 [20] and lncRNAs RMEL3 [21] and BANCR (BRAF-activated non-coding RNA) [22,23] activate the MAPK signaling pathway to induce cell proliferation even in the absence of GF. Indeed, up-regulation of miR-21 in melanoma is associated with worse prognosis [20,24,25]. This over-expression could be induced by environmental factors such as ultraviolet (UV) exposure that contribute to melanomagenesis [26,27]. Likewise, RMEL3 has been described as an enriched lncRNA in melanoma tumors [21,28], while BANCR stimulates MAPK pathways and the NOTCH cascade by sequestering miR-204 so that it is no longer able to repress NOTCH2 [22]. Meanwhile, miR-146a also participates in the regulation of this cascade by targeting LFNG (LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase) and the protein encoded by NUMB (numb homolog) [29,30]. Finally, tumor cell proliferation is also enhanced in melanoma cells by the lncRNA SAMMSON (survival associated mitochondrial melanoma-specific oncogenic non-coding RNA) that confers a growth advantage to melanoma cells. SAMMSON mainly localizes to the cytoplasm and to a lesser degree to mitochondria, where it interacts with p32, a protein implicated in mitochondrial metabolism [31]. However, SAMMSON functions and downstream growth enhancement mechanisms are largely unknown.
3. Non-Coding RNAs Increase Melanoma Cell Insensitivity to Growth Suppressor Signals
Another hallmark of tumor cell development lies in cell evasion of control by growth inhibitors, resulting in sustained proliferative signaling. The principal antigrowth factor, or growth inhibitor, is TGFβ (transforming growth factor beta). TGFβ and other antigrowth factors bind to transmembrane cell surface receptors coupled to intracellular signaling pathways (Figure 1), leading to regulation of the cell cycle clock specifically via cyclins and kinases that govern cell cycle transit through the G1 phase (Figure 2).
Several ncRNAs that target proteins of the cell cycle are significantly down-regulated in melanoma cells compared to normal melanocytes, whereas in the latter they act as tumor suppressor ncRNAs. The miRNA most differentially expressed between normal melanocytes and primary melanomas, miR-211 [32,33,34,35,36], targets TGFβ [35] to stop the advancement of the cell cycle [32,33,37]. Another TS-miR that is under-expressed in melanomas and in a variety of other cancers, miR-206, inhibits translation of cyclin-dependent kinases such as CDK4 (cyclin-dependent kinase 4), cyclin D1 or cyclin C to induce G1 arrest in noncancerous cells [38]; similar effects are also achieved by miR-let-7b and miR-193b [39,40,41]. Likewise, tumor suppressor proteins such as those encoded by CYLD (cylindromatosis) [42,43,44] are down-regulated by at least two miRNAs that are over-expressed in melanoma cells, miR-767 [45] and miR-186 [46].
Conversely, two ncRNAs acting as a pair, miR-221 and miR-222, have been reported to drive oncogenesis of melanomas and many types of malignancies through their dynamic roles in removing inhibitory controls of proliferation and progression [47,48,49]. For example, both miR-221 and miR-222 have been shown to target p27, another important cell cycle regulator that induces cell cycle arrest when it binds to and blocks the function of cyclin D1 [48,50]. Besides these miRNAs, the focally amplified lncRNA on chromosome 1 (FALEC) has been identified as an oncogenic lncRNA in melanoma [51] in addition to PVT1 [52,53,54] and UCA1 (urothelial cancer associated 1) [55]. FALEC affects p21 expression in melanoma by acting upon EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) [51], whereas PVT1 enhances cell proliferation through the regulation of cyclin D1 [52] and sequestration of miR-26b [53]. The interplay between lncRNAs and miRNAs is also illustrated by the activity of the ncRNA pair UCA1 and miR-507. The experimental depletion of UCA1 in melanoma cell lines induces G0/G1 cell cycle arrest with increased expression of miR-507, suggesting that miR-507 directly binds UCA1. Moreover, the transcription factor forkhead box protein M1 (FOXM1), an essential effector of G2/M phase transition, is also a direct target of miR-507. In melanoma, the elevated amount of UCA1 sequesters miR-507, rendering the latter unable to repress FOXM1 [55].
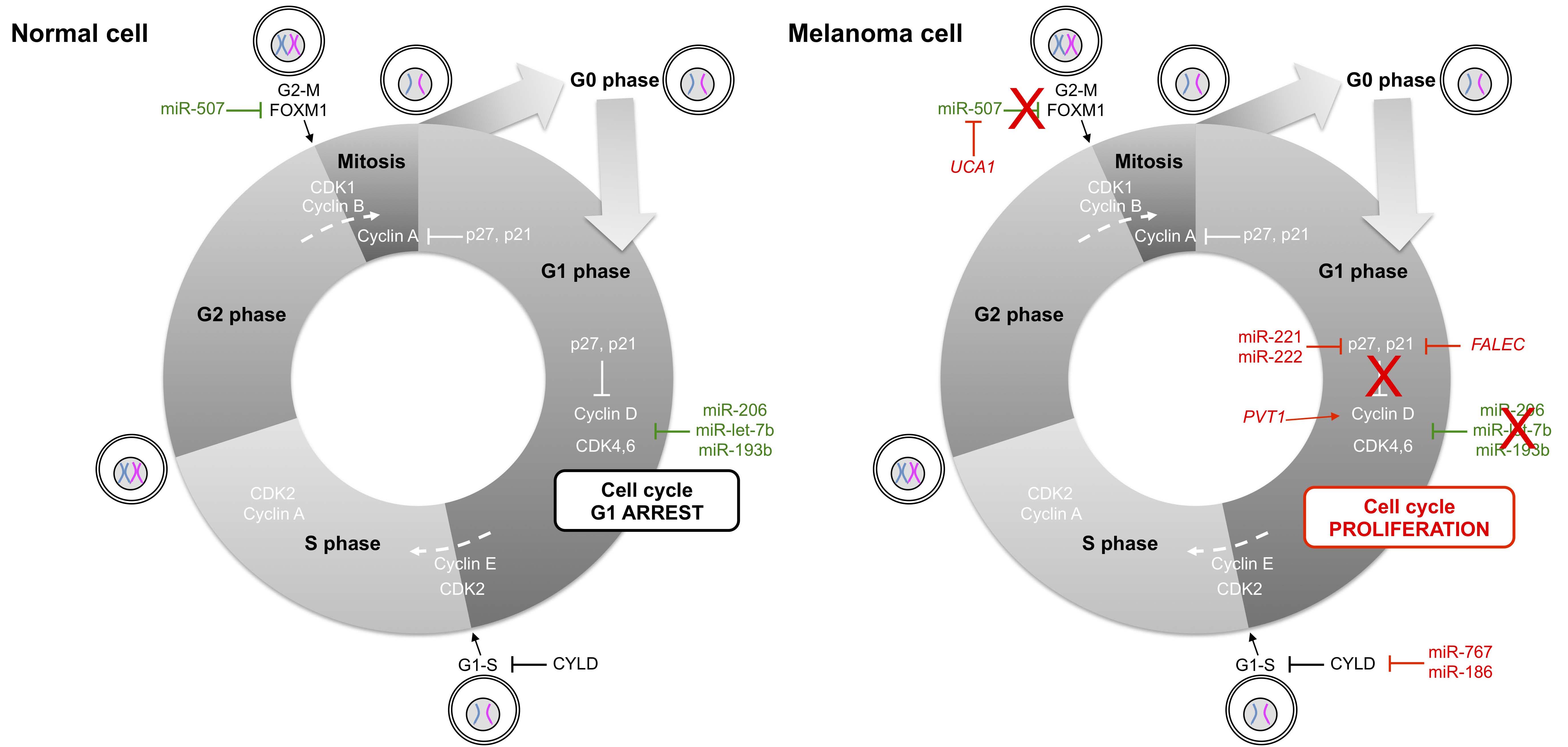
Figure 2 Non-coding RNAs in cell cycle control of normal and melanoma cells. Cyclin-dependent kinases (CDK) trigger the transition from G1 to S phase and from G2 to M phase by phosphorylating distinct sets of substrates. Cyclin D family members and CDK4,6 regulate events in the G1 phase, cyclin E/CDK2 triggers the S phase, cyclin A/CDK2 regulates the completion of the S phase and cyclin B/CDK1 is essential for driving cells into mitosis. In melanoma cells, aberrant expression of micro- and long ncRNAs activates mainly the cyclin D/CDK4,6 pair of ncRNAs, leading to progression from G1 to S phase. FOXM1: forkhead box M1; FALEC: focally amplified long non-coding RNA in epithelial cancer; PVT1: Pvt1 oncogene; CYLD: cylindromatosis; UCA1: urothelial cancer associated 1.
4. Non-Coding RNAs Enable Replicative Immortality by Circumvention of Senescence in Melanoma Cells
Uncontrolled cellular proliferation is a defining feature of cancer. In contrast, cellular senescence is a state of irreversible growth arrest which can be triggered by various stimuli such as telomere shortening, DNA damage, oxidative stress or oncogene activation (Figure 3). E2F family members are critical barriers to senescence induction that allow a cell to avoid senescence either through activation or inhibition of cell replication.
Some ncRNAs promote senescence in normal cells under selective stimuli, such as miR-205 [56] or miR-377 [57]. In normal cells, these ncRNAs target E2F family members and induce a senescence phenotype with elevated expression of p16INK4A, while both are down-regulated in melanoma cells [56,57]. UCA1, previously described in the regulation of the cell cycle, is also able to induce senescence by sequestering hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1) and by increasing p16INK4A mRNA stability to prevent its degradation [58]. Conversely, Montes et al. identified in melanoma patients a negative correlation between the lncRNA MIR31HG (MIR31 host gene) and p16INK4A [59]. Meanwhile, other studies suggest that p16INK4A could be also regulated by ANRIL (CDKN2B antisense RNA 1) through the recruitment of chromatin remodeling factors to its gene locus [60,61,62].
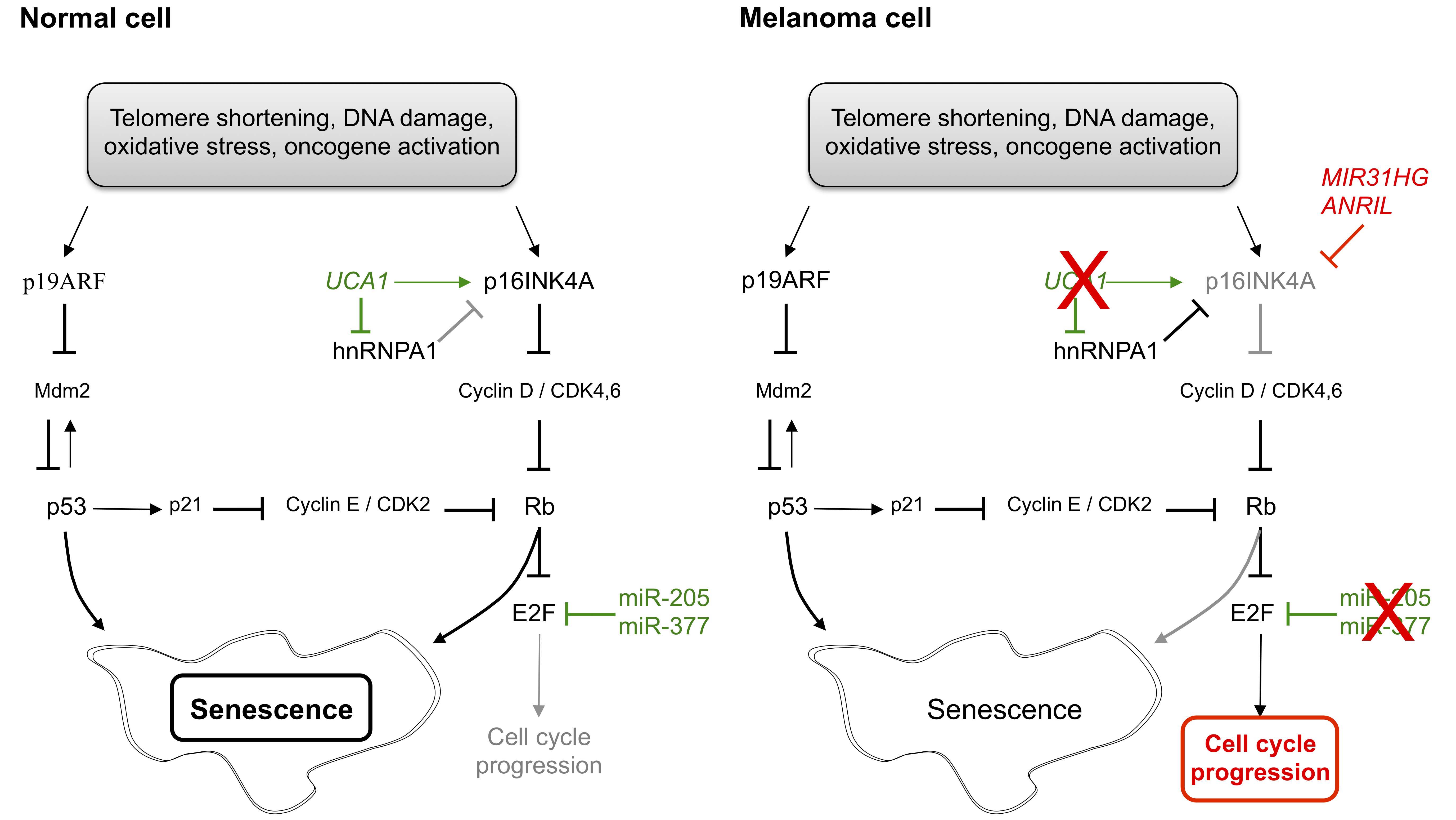
Figure 3 Regulation of cell senescence by non-coding RNAs in normal and melanoma cells. In normal cells responding to several signals, tumor suppressors p19ARF and p16INK4A are activated to prevent abnormal cell proliferation. In melanoma cells, p16INK4A is up-regulated by the expression of lncRNAs MIR31HG and ANRIL and upon down-regulation of miR-205, miR-377 and UCA1. p19ARF: cyclin-dependent kinase inhibitor 2A; Mdm2: MDM2 proto-oncogene, E3 ubiquitin protein ligase; UCA1: urothelial cancer associated 1; p16INK4A: cyclin-dependent kinase inhibitor 2A; hnRNPA1: heterogeneous nuclear ribonucleoprotein A1; CDK: cyclin-dependent kinase; Rb: retinoblastoma; MIR31HG: MIR31 host gene; ANRIL: CDKN2B antisense RNA 1.
5. Non-Coding RNAs Promote Melanoma Invasion and Metastasis
Melanoma cells are characterized by genetic instability and invasiveness through acquisition of migratory capacity. Several classes of proteins implicated as cell-cell adhesion molecules have been shown to be involved, such as immunoglobulin family members, calcium-dependent cadherin proteins or integrins, the latter of which link cells to the extracellular matrix. The metastatic capabilities of melanoma cancer cells also involve extracellular proteases such as MMP (matrix metallopeptidase) family members that are implicated in the breakdown of the extracellular matrix (Figure 4).
All of these pathways are regulated by ncRNAs. Indeed, miR-137 is down-regulated in melanoma cells [63,64,65] and represses transcription of the gene for E-cadherin by targeting the transcription factor TBX3 (T-box transcription factor 3) [66]. Meanwhile, integrin-dependent pathways that are regulated by miR-let-7a are also down-regulated in malignant melanoma cells [67]. Moreover, MMP proteins are regulated by the lncRNA GAS5 (growth arrest-specific 5) [68] and are also down-regulated in melanoma tissues [69], resulting in inhibition of their effects.
Conversely, numerous ncRNAs increase cell invasion by targeting the same pathways that function in cell migration. Thus, miR-21 (previously described in the first hallmark) targets TIMP3 (TIMP metallopeptidase inhibitor 3), a protein that inhibits MMP synthesis and activity [37,70]. MMP family members are also regulated by the lncRNA SLNCR1 (steroid receptor RNA activator 1-like non-coding RNA) through their interactions with Brn3a (brain-specific homeobox protein 3a) and AR (androgen receptor) transcription factors [71]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), first discovered in lung cancer, is also up-regulated in melanoma tumors [72]. MALAT1 similarly regulates extracellular proteases, while also inducing migration by sequestering miR-22 [73] and miR183 [74] through respective targeting of MMP14 and IGTB1 (Integrin β1). In addition to MALAT1 regulation, integrin pathways are also controlled by the ncRNA pair miR-221/miR-222 through enhancement of cellular migration via activation of the Akt/PI3K signaling pathway [50].
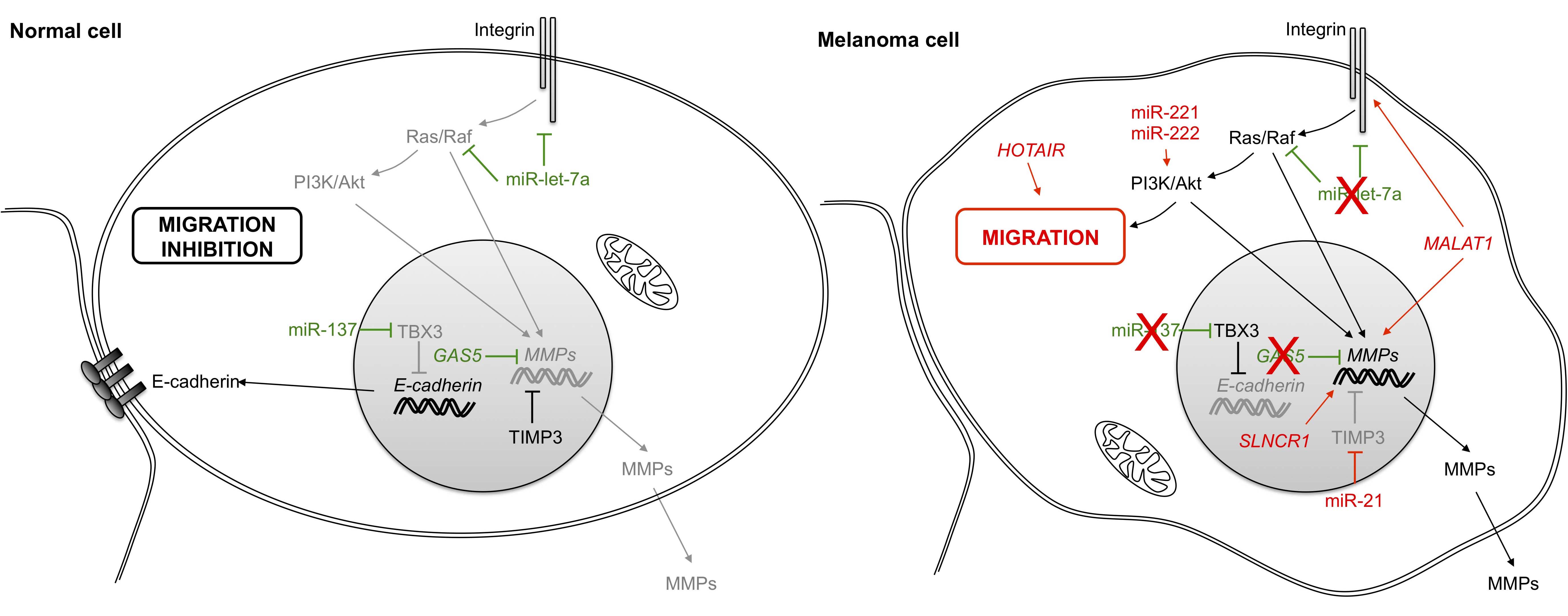
Figure 4 Role of non-coding RNAs in cell invasion and migration. In a melanoma cancer cell, integrin binding to ligands in the extracellular matrix triggers multiple signals leading to changes in gene expression and cell migration. The loss of the epithelial cell-cell adhesion molecule E-cadherin also participates by inducing cell migration. PI3K: phosphatidylinositol-4, 5-bisphosphate 3-kinase; Akt: v-akt murine thymoma viral oncogene homolog; TBX3: T-box 3; MMP: matrix metallopeptidase; GAS5: growth arrest-specific 5; TIMP3: TIMP metallopeptidase inhibitor 3; HOTAIR: HOX transcript antisense RNA; MALAT1: metastasis-associated lung adenocarcinoma transcript 1.
Overall, HOX transcript antisense RNA (HOTAIR) promotes melanoma cell invasion and migration by regulating the epithelial-to-mesenchymal transition (EMT) through sequestration of miR-152-3p [75]. HOTAIR can also regulate target gene expression by acting as a guide for PRC2 (polycomb repressive complex 2), leading to the epigenetic silencing of metastasis suppressor genes [76]. Notably, HOTAIR levels are elevated in melanoma cells and are associated with poor prognosis [75,77].
6. Non-Coding RNAs Induce Angiogenesis in Melanoma Tumors
Oxygen and nutrients supplied by the vasculature are crucial for tumor cell development and proliferation. The formation of new blood vessels is important for tumors to obtain an adequate supply of oxygen and nutrients and to evacuate metabolic waste and undergo metastatic processes (Figure 5). The principal regulator of this vascularization process is vascular endothelial growth factor (VEGF), a protein that is up-regulated by hypoxia and oncogenic signaling pathways.
Few non-coding RNAs have been described in melanoma angiogenesis; most of them target, either directly or indirectly, hypoxia inducible factor α (HIF1α) [78,79]. It has been suggested that because miR-21 is highly expressed in melanoma cells [25], it may target PTEN (phosphatase and tensin homolog) to activate MAPK signaling. In this way, PTEN may thereby enhance HIF1α expression and VEGF transcription, as observed in prostate cancer cells [79]. Similarly, Mir-21 may also enhance angiogenesis via its target TIMP3 [80], while a previous study showed that miR-424 enhances VEGF expression by down-regulating cullin 2 (CUL2), a protein that stabilizes HIFα within a sequestered cytoplasmic complex [81].
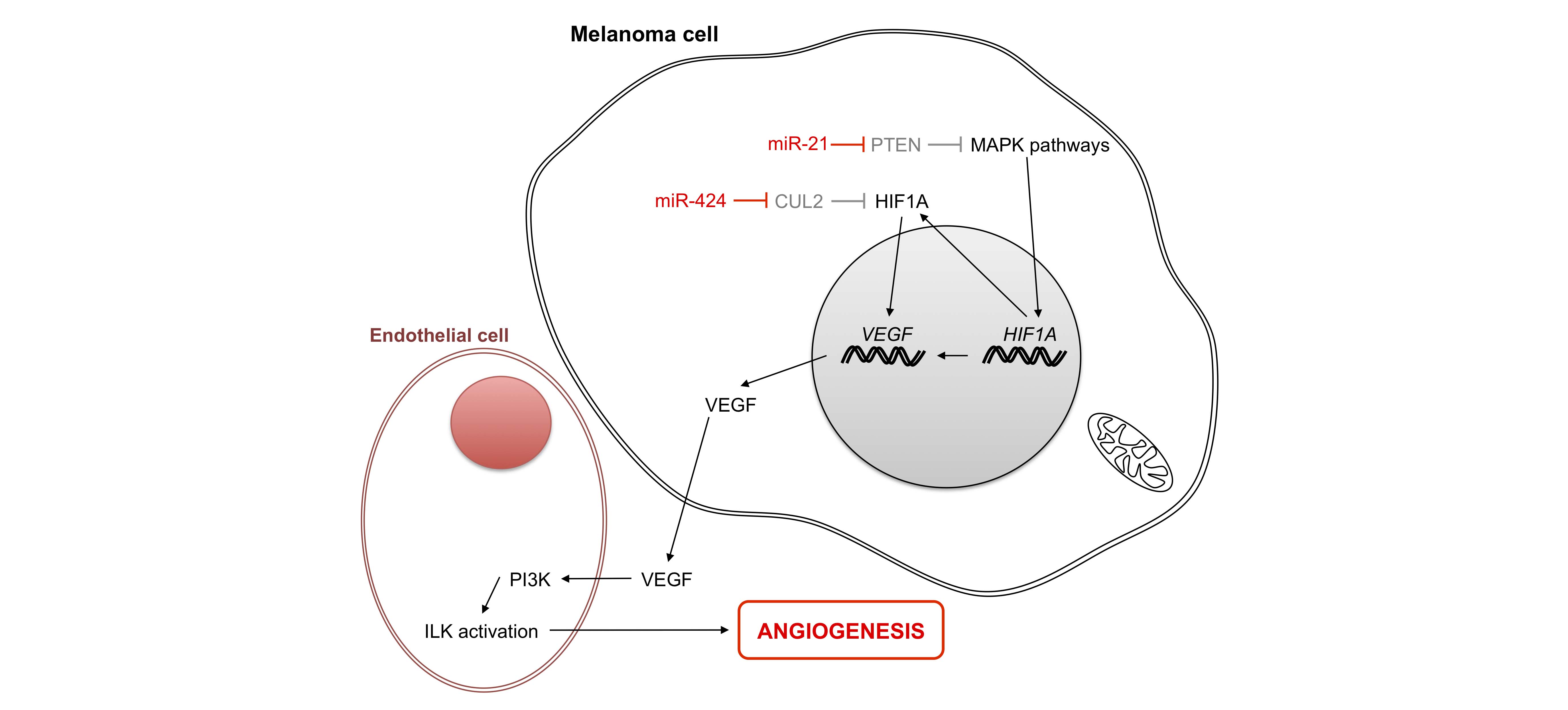
Figure 5 Role of micro-RNAs in angiogenesis. In the melanoma cell under normal oxygen conditions, HIF1α is degraded. Under hypoxia, HIF accumulates in the cytoplasm and moves to the nucleus to activate the production of proteins implicated in angiogenesis, including growth factor VEGF. Two micro-RNAs facilitate protein production by activating transcription of H1F1A and its accumulation in the cytoplasm. PTEN: phosphatase and tensin homolog; HIF1A: hypoxia inducible factor 1, alpha subunit; CUL2: cullin 2; VEGF: vascular endothelial growth factor A; ILK: integrin-linked kinase; PI3K: phosphatidylinositol-4, 5-bisphosphate 3-kinase.
7. Non-Coding RNAs Allow Evasion of Programmed Cell Death of Melanoma Cells
The ability of tumor cells to proliferate also stems from their resistance to programmed cell death. Apoptosis is dependent upon sensors which control the intra- and extracellular microenvironment, such as cell surface receptors and effectors which trigger cell death. Intracellular signals of apoptosis converge to act within mitochondria, where Bcl-2 family members play their anti- or pro-apoptotic roles through preventing or promoting the release of cytochrome C, respectively, with the latter leading to the activation of effector caspases. At least three TS-miRNAs, miR-219 [82], miR-15b [83,84] and miR-181a [85], enhance apoptosis by targeting Bcl-2. All of these TS-miRNAs are down-regulated in melanoma cells.
Overall, the lncRNA GAS5 regulates apoptosis in melanoma via its interaction with the DNA binding domain of GRs (glucocorticoid receptors) by preventing binding of GRs to glucocorticoid response elements (GREs) in the genome. This suppresses the induction of several response genes implicated in apoptosis [86].Finally, SPRY4-IT1 (SPRY4-IT1 intronic transcript 1) is over-represented in melanoma cells [87,88] and may block apoptosis by modulating lipid metabolism and lipotoxicity [89].
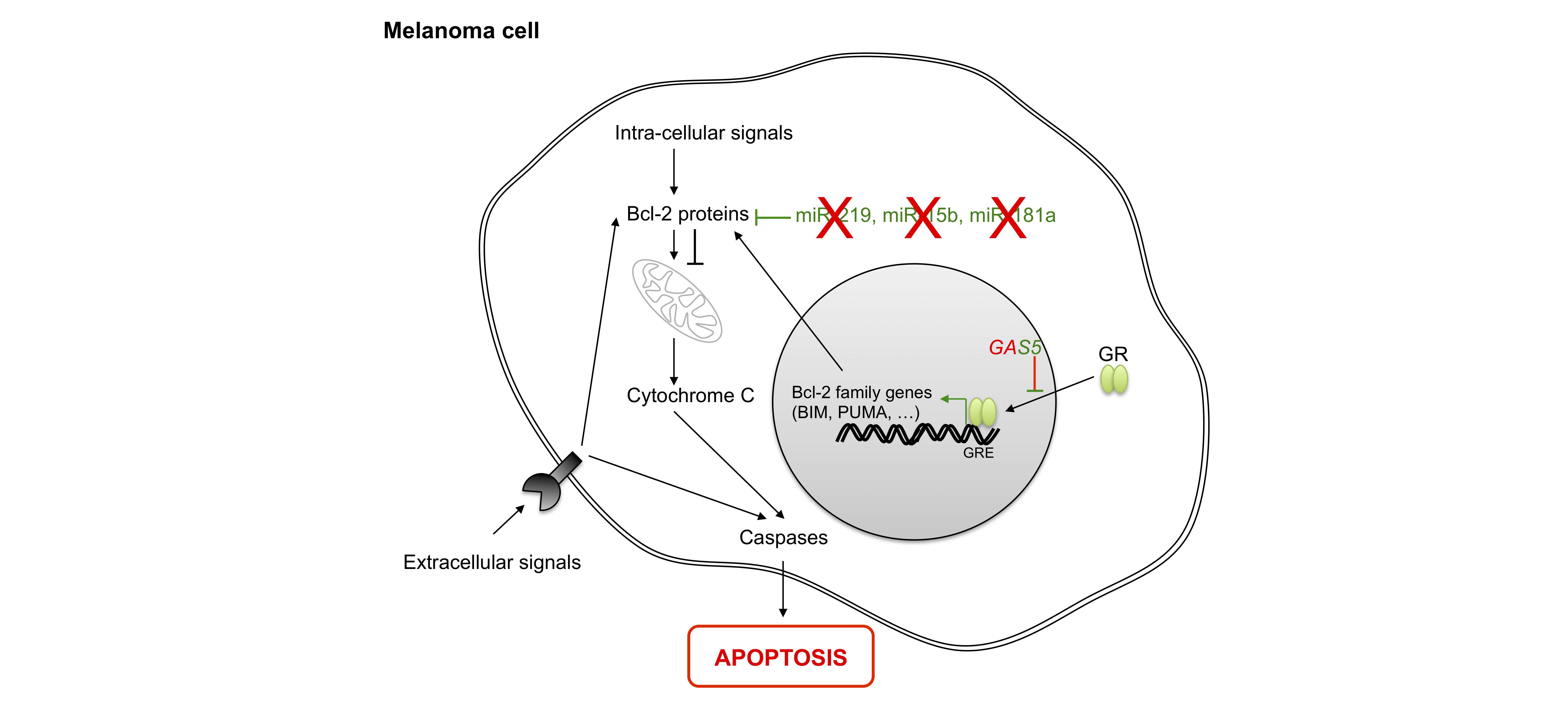
Figure 6 Role of non-coding RNAs in apoptosis. Apoptosis is induced by the activation of death receptors (extracellular signals) or intracellular signals, leading to the activation of Bcl-2 family members. Next, cytochrome c release activates effector caspases. In the nucleus, most Bcl-2 family genes are regulated by the binding of glucocorticoid receptor (GR) to their DNA sequences. BIM: BCL2-like 11; PUMA: BCL2 binding component 3; GAS5: growth arrest-specific 5; Bcl-2: B-cell CLL/lymphoma 2.
8. Conclusions
Among more than 3,000 miRNAs [90] and 24,793 annotated lncRNAs that have been identified within the human genome [91], only a few show deregulations in cutaneous melanoma (Table 1). Nevertheless, we are only at the beginning of our understanding and identification of ncRNAs; recent computational analyses using high-throughput expression data, including microarrays and RNA sequencing, have helped us identify deregulated mRNAs, lncRNAs and pseudogenes in melanoma [92,93]. Consequently, the growing body of evidence strongly suggests a role of pseudogene transcription as a source of endogenous siRNAs that either bind to their cognate wild-type (WT) genes or act as sponges by binding miRNAs targeting their WT genes. The emerging picture underscores the functional interdependence of coding and non-coding RNAs [94,95] whereby all perturbations in these interactions have widespread consequences that can lead to the malignant phenotypes of cancer cells.
Table 1 Non-coding RNAs deregulated in melanoma
|
Name |
Functions in tumor |
Targets |
Up/down-regulated in melanoma |
|
lncRNA |
|
|
|
|
ANRIL |
Senescence |
p16INK4A |
Up |
|
BANCR |
Proliferation |
mir-204 |
Up |
|
FALEC |
Cell cycle |
p21 |
Up |
|
GAS5 |
Invasion / Metastasis / Apoptosis |
MMP, GR |
Up |
|
HOTAIR |
Invasion / Metastasis |
PRC2 |
Up |
|
MALAT1 |
Invasion / Metastasis |
miR-22, miR-183 |
Up |
|
MIR31HG |
Senescence |
p16INK4A |
Up |
|
PVT1 |
Cell cycle |
Cyclin D1, miR-26b |
Up |
|
RMEL3 |
Proliferation |
|
Up |
|
SAMMSON |
Proliferation |
p32 |
Up |
|
SLNCR1 |
Invasion / Metastasis |
Brn3a, AR |
Up |
|
SPRY4-IT1 |
Apoptosis |
|
Up |
|
UCA1 |
Cell cycle / Senescence |
miR-507, hnRNPA1 |
Up |
|
miRNA |
|
|
|
|
miR-137 |
Invasion / Metastasis |
TBX3 |
Down |
|
miR-146a |
Proliferation |
LFNG, NUMB |
Up |
|
miR-15b |
Apoptosis |
Bcl-2 family members |
Down |
|
miR-181a |
Apoptosis |
Bcl-2 family members |
Down |
|
miR-186 |
Cell cycle |
CYLD |
Up |
|
miR-193b |
Cell cycle |
CDK4, Cyclin D1, Cyclin C |
Down |
|
miR-204 |
Proliferation |
|
Down |
|
miR-205 |
Senescence |
E2F members |
Down |
|
miR-206 |
Cell cycle |
CDK4, Cyclin D1, Cyclin C |
Down |
|
mir-21 |
Proliferation / Invasion / Metastasis / Angiogenesis |
TIMP3, PTEN |
Up |
|
miR-211 |
Growth |
TGFB |
Down |
|
miR-219 |
Apoptosis |
Bcl-2 family members |
Down |
|
miR-221 / 222 |
Cell cycle / Invasion / Metastasis |
p27, Akt/PI3K |
Up |
|
miR-31 |
Proliferation |
MET, RAB27, NIK |
Down |
|
miR-377 |
Senescence |
E2F members |
Down |
|
miR-424 |
Angiogenesis |
CUL2 |
Up |
|
miR-767 |
Cell cycle |
CYLD |
Up |
|
miR-let-7a |
Invasion / Metastasis |
Integrin |
Down |
|
miR-let-7b |
Cell cycle |
CDK4, Cyclin D1, Cyclin C |
Down |
A more complete understanding of the cellular biology of ncRNAs, notably lncRNAs, is crucial to improving early diagnosis, prognosis and treatment. The latter, of course, first requires detailed knowledge about melanoma-specific lncRNAs functions. However, therapeutic modulation of cancer- and tissue-specific expression of lncRNAs could have major advantages over other therapeutics.
We also must consider the timing of action of ncRNAs, since many of the studies described in this review only investigated ncRNAs present at the time of diagnosis. Further studies need to be done to determine the variation in ncRNAs levels throughout the disease course to further evaluate their relevance to survival and treatment.
To conclude, the study of ncRNA effects on melanoma is a very attractive undertaking, since they participate in numerous cancer processes. Notably, because ncRNAs could be released into the bloodstream, they may serve as non-invasive biomarkers of melanoma activity.
Author Contributions
VV and DF wrote the manuscript and designed figures.
Competing Interests
The authors have declared that no competing interests exist.
References
- Abdel-Rahman O. Evaluation of the eighth American Joint Committee on Cancer staging system for malignant melanoma of the skin. Future Oncol. 2018; 14: 471-481. [CrossRef] [Google scholar] [PubMed]
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016; 66: 271-289. [CrossRef] [Google scholar] [PubMed]
- Kuske M, Rauschenberg R, Garzarolli M, Meredyth-Stewart M, Beissert S, Troost EGC, et al. Melanoma Brain Metastases: Local Therapies, Targeted Therapies, Immune Checkpoint Inhibitors and Their Combinations-Chances and Challenges. Am J Clin Dermatol. 2018. [CrossRef] [Google scholar] [PubMed]
- Najem A, Krayem M, Perdrix A, Kerger J, Awada A, Journe F, et al. New Drug Combination Strategies in Melanoma: Current Status and Future Directions. Anticancer Res. 2017; 37: 5941-5953. [Google scholar]
- Marconcini R, Spagnolo F, Stucci LS, Ribero S, Marra E, Rosa F, et al. Current status and perspectives in immunotherapy for metastatic melanoma. Oncotarget. 2018; 9: 12452-12470. [CrossRef] [Google scholar] [PubMed]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012; 338: 1435-1439. [CrossRef] [Google scholar] [PubMed]
- Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol. 1955; 1: 59-68. [CrossRef] [Google scholar] [PubMed]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958; 231: 241-257. [Google scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014; 15: 423-437. [CrossRef] [Google scholar] [PubMed]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011; 331: 550-553. [CrossRef] [Google scholar] [PubMed]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002; 21: 4663-4670. [CrossRef] [Google scholar] [PubMed]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19: 92-105. [CrossRef] [Google scholar] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006; 6: 259-269. [CrossRef] [Google scholar] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120 : 15-20. [CrossRef] [Google scholar] [PubMed]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22: 1775-1789. [CrossRef] [Google scholar] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000; 100: 57-70. [CrossRef] [Google scholar] [PubMed]
- Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002; 12: 9-18. [CrossRef] [Google scholar] [PubMed]
- Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012; 3: 1011-1025. [CrossRef] [Google scholar] [PubMed]
- Poliseno L, Haimovic A, Segura MF, Hanniford D, Christos PJ, Darvishian F, et al. Histology-specific microRNA alterations in melanoma. J Invest Dermatol. 2012; 132: 1860-1868. [CrossRef] [Google scholar] [PubMed]
- Jiang L, Lv X, Li J, Li J, Li X, Li W, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012; 114: 582-588. [CrossRef] [Google scholar] [PubMed]
- Goedert L, Pereira CG, Roszik J, Placa JR, Cardoso C, Chen G, et al. RMEL3, a novel BRAFV600E-associated long noncoding RNA, is required for MAPK and PI3K signaling in melanoma. Oncotarget. 2016; 7: 36711-36718. [CrossRef] [Google scholar] [PubMed]
- Cai B, Zheng Y, Ma S, Xing Q, Wang X, Yang B, et al. BANCR contributes to the growth and invasion of melanoma by functioning as a competing endogenous RNA to upregulate Notch2 expression by sponging miR204. Int J Oncol. 2017; 51: 1941-1951. [CrossRef] [Google scholar] [PubMed]
- Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, et al. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014; 9: e100893. [CrossRef] [Google scholar] [PubMed]
- Grignol V, Fairchild ET, Zimmerer JM, Lesinski GB, Walker MJ, Magro CM, et al. miR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br J Cancer. 2011; 105: 1023-1029. [CrossRef] [Google scholar] [PubMed]
- Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, Kapp A, et al. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp Dermatol. 2012; 21: 509-514. [CrossRef] [Google scholar] [PubMed]
- Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011; 286: 39172-39178. [CrossRef] [Google scholar] [PubMed]
- Melnik BC. MiR-21: an environmental driver of malignant melanoma? J Transl Med. 2015; 13: 202. [CrossRef] [Google scholar] [PubMed]
- Sousa JF, Torrieri R, Silva RR, Pereira CG, Valente V, Torrieri E, et al. Novel primate-specific genes, RMEL 1, 2 and 3, with highly restricted expression in melanoma, assessed by new data mining tool. PLoS One. 2010; 5: e13510. [CrossRef] [Google scholar] [PubMed]
- Forloni M, Dogra SK, Dong Y, Conte D, Jr., Ou J, Zhu LJ, et al. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife. 2014; 3: e01460. [CrossRef] [Google scholar] [PubMed]
- Raimo M, Orso F, Grassi E, Cimino D, Penna E, De Pitta C, et al. miR-146a Exerts Differential Effects on Melanoma Growth and Metastatization. Mol Cancer Res. 2016; 14: 548-562. [CrossRef] [Google scholar] [PubMed]
- Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016; 531: 518-522. [CrossRef] [Google scholar] [PubMed]
- Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys. 2014; 563: 60-70. [CrossRef] [Google scholar] [PubMed]
- Weiss SA, Hanniford D, Hernando E, Osman I. Revisiting determinants of prognosis in cutaneous melanoma. Cancer. 2015; 121: 4108-4123. [CrossRef] [Google scholar] [PubMed]
- Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma R. 2011; 24: 525-537. [CrossRef] [Google scholar] [PubMed]
- Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010; 40: 841-849. [CrossRef] [Google scholar] [PubMed]
- Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One. 2010; 5: e13779. [CrossRef] [Google scholar] [PubMed]
- Latchana N, Ganju A, Howard JH, Carson WE, 3rd. MicroRNA dysregulation in melanoma. Surg Oncol. 2016; 25: 184-189. [CrossRef] [Google scholar] [PubMed]
- Georgantas RW, 3rd, Streicher K, Luo X, Greenlees L, Zhu W, Liu Z, et al. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma R. 2014; 27: 275-286. [CrossRef] [Google scholar] [PubMed]
- Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010; 176: 2520-2529. [CrossRef] [Google scholar] [PubMed]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008; 18: 549-557. [CrossRef] [Google scholar] [PubMed]
- Xu D, Tan J, Zhou M, Jiang B, Xie H, Nie X, et al. Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett. 2012; 4: 941-946. [CrossRef] [Google scholar] [PubMed]
- Pannem RR, Dorn C, Ahlqvist K, Bosserhoff AK, Hellerbrand C, Massoumi R. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis. 2014; 35: 461-468. [CrossRef] [Google scholar] [PubMed]
- Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, et al. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-kappaB signaling. J Transl Med. 2014; 12: 33. [CrossRef] [Google scholar] [PubMed]
- Yang Y, Zhou J. CYLD - a deubiquitylase that acts to fine-tune microtubule properties and functions. J Cell Sci. 2016; 129: 2289-2295. [CrossRef] [Google scholar] [PubMed]
- Zhang K, Guo L. MiR-767 promoted cell proliferation in human melanoma by suppressing CYLD expression. Gene. 2018; 641: 272-278. [CrossRef] [Google scholar] [PubMed]
- Qiu H, Yuan S, Lu X. miR-186 suppressed CYLD expression and promoted cell proliferation in human melanoma. Oncol Lett. 2016; 12: 2301-2306. [CrossRef] [Google scholar] [PubMed]
- Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012; 12: 27-33. [CrossRef] [Google scholar] [PubMed]
- Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008; 68: 2745-2754. [CrossRef] [Google scholar] [PubMed]
- Errico MC, Felicetti F, Bottero L, Mattia G, Boe A, Felli N, et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int J Cancer. 2013; 133: 879-892. [CrossRef] [Google scholar] [PubMed]
- Felicetti F, De Feo A, Coscia C, Puglisi R, Pedini F, Pasquini L, et al. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J Transl Med. 2016; 14: 56. [CrossRef] [Google scholar] [PubMed]
- Ni N, Song H, Wang X, Xu X, Jiang Y, Sun J. Up-regulation of long noncoding RNA FALEC predicts poor prognosis and promotes melanoma cell proliferation through epigenetically silencing p21. Biomed Pharmacother. 2017; 96: 1371-1379. [CrossRef] [Google scholar] [PubMed]
- Chen L, Ma D, Li Y, Li X, Zhao L, Zhang J, et al. Effect of long non-coding RNA PVT1 on cell proliferation and migration in melanoma. Int J Mol Med. 2018; 41: 1275-1282. [Google scholar]
- Wang BJ, Ding HW, Ma GA. Long Noncoding RNA PVT1 Promotes Melanoma Progression Via Endogenous Sponging MiR-26b. Oncol Res. 2017. [CrossRef] [Google scholar] [PubMed]
- Chen X, Gao G, Liu S, Yu L, Yan D, Yao X, et al. Long Noncoding RNA PVT1 as a Novel Diagnostic Biomarker and Therapeutic Target for Melanoma. Biomed Res Int. 2017; 2017: 7038579. [CrossRef] [Google scholar] [PubMed]
- Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016; 33: 88. [CrossRef] [Google scholar] [PubMed]
- Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011; 286: 16606-16614. [CrossRef] [Google scholar] [PubMed]
- Zehavi L, Schayek H, Jacob-Hirsch J, Sidi Y, Leibowitz-Amit R, Avni D. MiR-377 targets E2F3 and alters the NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol Cancer. 2015; 14: 68. [CrossRef] [Google scholar] [PubMed]
- Kumar PP, Emechebe U, Smith R, Franklin S, Moore B, Yandell M, et al. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. Elife. 2014; 3. [CrossRef] [Google scholar] [PubMed]
- Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015; 6: 6967. [CrossRef] [Google scholar] [PubMed]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010; 38: 662-674. [CrossRef] [Google scholar] [PubMed]
- Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010; 6: e1000899. [CrossRef] [Google scholar] [PubMed]
- Xu S, Wang H, Pan H, Shi Y, Li T, Ge S, et al. ANRIL lncRNA triggers efficient therapeutic efficacy by reprogramming the aberrant INK4-hub in melanoma. Cancer Lett. 2016; 381: 41-48. [CrossRef] [Google scholar] [PubMed]
- Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013; 133: 768-775. [CrossRef] [Google scholar] [PubMed]
- Bennett PE, Bemis L, Norris DA, Shellman YG. miR in melanoma development: miRNAs and acquired hallmarks of cancer in melanoma. Physiol Genomics. 2013; 45: 1049-1059. [CrossRef] [Google scholar] [PubMed]
- Li N. Low Expression of Mir-137 Predicts Poor Prognosis in Cutaneous Melanoma Patients. Med Sci Monit. 2016; 22: 140-144. [CrossRef] [Google scholar] [PubMed]
- Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008; 68: 7872-7881. [CrossRef] [Google scholar] [PubMed]
- Muller DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008; 27: 6698-6706. [CrossRef] [Google scholar] [PubMed]
- Chen L, Yang H, Xiao Y, Tang X, Li Y, Han Q, et al. Lentiviral-mediated overexpression of long non-coding RNA GAS5 reduces invasion by mediating MMP2 expression and activity in human melanoma cells. Int J Oncol. 2016; 48: 1509-1518. [CrossRef] [Google scholar] [PubMed]
- Bian D, Shi W, Shao Y, Li P, Song G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am J Transl Res. 2017; 9: 1509-1520. [Google scholar]
- Martin del Campo SE, Latchana N, Levine KM, Grignol VP, Fairchild ET, Jaime-Ramirez AC, et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: in vivo effects of MiR-21 inhibitor. PLoS One. 2015; 10: e0115919. [CrossRef] [Google scholar] [PubMed]
- Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, et al. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016; 15: 2025-2037. [CrossRef] [Google scholar] [PubMed]
- Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014; 24: 335-341. [CrossRef] [Google scholar] [PubMed]
- Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, et al. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016; 7: 63901-63912. [CrossRef] [Google scholar] [PubMed]
- Sun Y, Cheng H, Wang G, Yu G, Zhang D, Wang Y, et al. Deregulation of miR-183 promotes melanoma development via lncRNA MALAT1 regulation and ITGB1 signal activation. Oncotarget. 2017; 8: 3509-3518. [CrossRef] [Google scholar] [PubMed]
- Luan W, Li R, Liu L, Ni X, Shi Y, Xia Y, et al. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote malignant melanoma progression by sponging miR-152-3p. Oncotarget. 2017; 8: 85401-85414. [CrossRef] [Google scholar] [PubMed]
- Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013; 52: 9519-9527. [CrossRef] [Google scholar] [PubMed]
- Cantile M, Scognamiglio G, Marra L, Aquino G, Botti C, Falcone MR, et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J Cell Physiol. 2017; 232: 3422-3432. [CrossRef] [Google scholar] [PubMed]
- Hwang HW, Baxter LL, Loftus SK, Cronin JC, Trivedi NS, Borate B, et al. Distinct microRNA expression signatures are associated with melanoma subtypes and are regulated by HIF1A. Pigment Cell Melanoma Res. 2014; 27: 777-787. [CrossRef] [Google scholar] [PubMed]
- Liu LZ, Li C, Chen Q, Jing Y, Carpenter R, Jiang Y, et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS One. 2011; 6: e19139. [CrossRef] [Google scholar] [PubMed]
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med. 2003; 9: 407-415. [CrossRef] [Google scholar] [PubMed]
- Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010; 120: 4141-4154. [CrossRef] [Google scholar] [PubMed]
- Long J, Menggen Q, Wuren Q, Shi Q, Pi X. MiR-219-5p Inhibits the Growth and Metastasis of Malignant Melanoma by Targeting BCL-2. Biomed Res Int. 2017; 2017: 9032502. [CrossRef] [Google scholar] [PubMed]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008; 123: 372-379. [CrossRef] [Google scholar] [PubMed]
- Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med. 2012; 10: 155. [CrossRef] [Google scholar] [PubMed]
- Du M, Zhang Z, Gao T. Piceatannol induced apoptosis through up-regulation of microRNA-181a in melanoma cells. Biol Res. 2017; 50: 36. [CrossRef] [Google scholar] [PubMed]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010; 3: ra8. [CrossRef] [Google scholar] [PubMed]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011; 71: 3852-3862. [CrossRef] [Google scholar] [PubMed]
- Liu T, Shen SK, Xiong JG, Xu Y, Zhang HQ, Liu HJ, et al. Clinical significance of long noncoding RNA SPRY4-IT1 in melanoma patients. FEBS Open Bio. 2016; 6: 147-154. [CrossRef] [Google scholar] [PubMed]
- Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014; 5: 8959-8969. [CrossRef] [Google scholar] [PubMed]
- Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA. 2015; 112: E1106-1115. [CrossRef] [Google scholar] [PubMed]
- Ning Q, Li Y, Wang Z, Zhou S, Sun H, Yu G. The Evolution and Expression Pattern of Human Overlapping lncRNA and Protein-coding Gene Pairs. Sci Rep. 2017; 7: 42775. [CrossRef] [Google scholar] [PubMed]
- Capobianco E, Valdes C, Sarti S, Jiang Z, Poliseno L, Tsinoremas NF. Ensemble Modeling Approach Targeting Heterogeneous RNA-Seq data: Application to Melanoma Pseudogenes. Sci Rep. 2017; 7: 17344. [CrossRef] [Google scholar] [PubMed]
- Zhao H, Li Y, Wang S, Yang Y, Wang J, Ruan X, et al. Whole transcriptome RNA-seq analysis: tumorigenesis and metastasis of melanoma. Gene. 2014; 548: 234-243. [CrossRef] [Google scholar] [PubMed]
- Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012; 149: 1622-1634. [CrossRef] [Google scholar] [PubMed]
- Milligan MJ, Harvey E, Yu A, Morgan AL, Smith DL, Zhang E, et al. Global Intersection of Long Non-Coding RNAs with Processed and Unprocessed Pseudogenes in the Human Genome. Front Genet. 2016; 7: 26. [CrossRef] [Google scholar] [PubMed]



