Pneumocystis jirovecii Pneumonia: Current Advances in Laboratory Diagnosis
Ana Luísa Tomás † ![]() , Olga Matos †, *
, Olga Matos †, * ![]()
Medical Parasitology Unit, Group of Opportunistic Protozoa/HIV and Other Protozoa, Global Health and Tropical Medicine, Instituto de Higiene e Medicina Tropical, Universidade Nova de Lisboa, Rua da Junqueira 100, Lisboa, Portugal
† These authors contributed equally to this work.
Received: August 21, 2018 | Accepted: November 05, 2018 | Published: November 13, 2018
OBM Genetics 2018, Volume 2, Issue 4 doi: 10.21926/obm.genet.1804049
Academic Editors: Andrés Moya, Enrique J. Calderón and Luis Delaye
Special Issue: Pneumocystis: A Model of Adaptive Coevolution
Recommended citation: Tomás AL, Matos O. Pneumocystis jirovecii Pneumonia: Current Advances in Laboratory Diagnosis. OBM Genetics 2018;2(4):049; doi:10.21926/obm.genet.1804049.
© 2018 by the authors. This is an open access article distributed under the conditions of the Creative Commons by Attribution License, which permits unrestricted use, distribution, and reproduction in any medium or format, provided the original work is correctly cited.
Abstract
Pneumocystis jirovecii pneumonia (PcP) remains a major cause of respiratory illness among immunocompromised patients. PcP is difficult to diagnose, in particular in non-HIV-infected patients, due to the lack of associated specific clinical data. Since P. jirovecii could not be cultivated for many years, microscopic visualization of cystic or trophic forms in respiratory specimens based on cytochemical or immunofluorescence staining are the standard procedure to identify this fungus. Polymerase chain reaction (PCR)-based methodologies have been developed to overcome the low sensitivity of microscopy in respiratory specimens, especially those with low fungal load and in non-HIV-infected patients. Real-time quantitative PCR is the only format suitable for a quantitative diagnosis, and these results have been used to differentiate PcP active disease (high fungal load) from carriage/colonization (low fungal load). However, its use is inconclusive with limited results in intermediate fungal loads. New strategies based on measurement of blood biomarkers may be a viable alternative to perform PcP diagnosis non-invasively. Several studies explored the usefulness of candidate serum biomarkers, such as (1-3)-β-D-Glucan, Krebs von den Lungen-6 antigen, lactate dehydrogenase, and S-adenosylmethionine, with the former presenting the most promising results. More recently, approaches based on the detection of specific anti-P. jirovecii antibodies in patients’ sera are showing encouraging results that could enable a faster and inexpensive screening and diagnosis of this opportunistic infectious disease, helping to improve therapeutic interventions, disease control, and provide retrenchment to healthcare systems.
Keywords
Pneumocystis jirovecii; pneumonia; laboratory diagnosis; current methods; new alternatives
1. Pneumocystis Pneumonia Infection
Pneumocystis jirovecii (previously called Pneumocystis carinii f. sp. hominis) is an opportunistic fungus of ubiquitous distribution with specificity restricted to humans. This pathogen is capable of causing fatal interstitial pneumonia (PcP) in immunocompromised hosts, especially in those infected by the human immunodeficiency virus (HIV), but also in patients who are undergoing immunosuppressive treatments related to other pathologies. This disease is also emerging as a co-morbidity factor associated with chronic diseases, such as chronic obstructive pulmonary disorder (COPD), and can cause asymptomatic infection in immunocompetent persons [1,2,3]. Pneumocystis infections are, as a rule, confined to the lungs. However, infections in other organs or tissues have been reported [4].
During the last 30 years, PcP proved to be an extremely important disease associated with HIV infection and the acquired immune deficiency syndrome (AIDS) epidemic. During the first years of the HIV/AIDS epidemic, PcP functioned as the main indicator associated with HIV infection; in recent years, it still plays a relevant role in the clinical picture of HIV-infected patients, since recent reports point to PcP as the most common AIDS-defining illness in Europe in 2016. This disease was recorded in 20.0% of the cases, followed by pulmonary and/or extra-pulmonary tuberculosis (15%), oesophageal candidiasis (11%), and wasting syndrome due to HIV (10%). This demonstrates that even in the era of combination antiretroviral therapy (cART), PcP remains a significant cause of morbidity and mortality in patients with HIV/AIDS in developed countries [5]. In developing countries, the scenario is even worse because in addition to the large number of HIV-infected patients, access to cART, PcP diagnosis, and prophylaxis is limited due to lack of resources and expertise [6,7,8,9].
Therefore, the control and prevention of this disease is still an area that requires much attention from a public health point of view in all countries. One way to meet this need is by improving the tools for an early and accurate diagnosis of Pneumocystis pneumonia.
2. Clinical and Laboratory Assessment of PcP
PcP does not present pathognomonic clinical, radiologic or gasometric findings. Therefore, the diagnosis of this disease depends on a clinical assessment based on non-specific clinical manifestations, pulmonary function, arterial blood gas and radiological testing, plus non-specific and specific laboratory tests. Nevertheless, PcP presentation depends on the underlying disease and usually the clinical and laboratory findings are less severe in patients immunocompromised by other pathologies rather than HIV infection [10].
The non-specific clinical manifestations of a patient with PcP include fever, non-productive cough and dyspnea. The most common radiological presentation of this pathology is a pattern of bilateral interstitial pneumonia, but may vary depending on the degree of immune deficiency, the presence of other concomitant infections, or the use of pentamidine in the prophylaxis of PcP. Blood gas analysis shows hypoxemia that worsens with exercise and a partial pressure of oxygen (PaO2) in peripheral blood ≤ 9.3 kPa (70 mmHg) is indicative of PcP [1,11]. In terms of non-specific laboratory tests, the measurement of increased levels of lactate dehydrogenase (LDH) can be used as a prognostic tool and to assess response to therapy [12]. Since the risk of developing the disease increases with a CD4+ T cell count ≤ 200/mm3 in all patients [1,13,14], this analysis is crucial not only for the diagnosis, but also for the prevention and control of PcP. In spite of the usefulness of all these clinical and laboratory data, a definitive diagnosis still depends on the detection of P. jirovecii in the affected tissues, which is only possible with specific laboratory methodologies.
In contrast to most pathogenic microorganisms, the absence of a sustained, stable, and reproducible P. jirovecii culture medium has been a significant limitation for the disease diagnosis [15]. Although a promising culture system to propagate P. jirovecii in vitro was developed in 2014 [16], it still needs to be validated since some authors were unable to reproduce the results obtained in 2014 [17]. Thus, the isolation, cultivation, and propagation of this fungus remains a challenge for diagnostic purposes.
Meanwhile, the evolution and improvement of classical diagnostic methods (such as cytochemical or immunofluorescent staining and detection of DNA of the organism by molecular techniques) have enabled an improvement in the detection of P. jirovecii, mainly in respiratory specimens. However, the successful diagnosis of PcP based on these methods is dependent on the resources and expertise of the laboratory team, as well as the type of biological specimen analysed. Therefore, finding simple methods and minimally invasive specimens, such as blood for rapid and effective detection of the causative agent, has become an urgent need. This need is especially critical in low-to-middle income countries, where it is difficult to implement classical laboratory diagnostic techniques and the appropriate methods for respiratory specimen collection, since both require specialized personnel, expensive equipment, and structures that are not readily available, in most cases [9,18]. This urgency leads to developing new strategies and new approaches for PcP diagnosis.
2.1. Classical Methods for PcP Laboratorial Diagnosis
The classical diagnosis of PcP is based on the identification or detection of P. jirovecii in the affected tissues by cytochemical staining, immunofluorescent staining with monoclonal antibodies (IFI-MAb), and/or the detection of Pneumocystis DNA by molecular techniques. The most important characteristics of the commonly applied and studied laboratory diagnostic methods for PcP classical diagnosis are summarized in Table 1.
Table 1 Characteristics of the classical laboratory methods for PcP diagnosis (adapted from [26]).

Biological specimens. Specimens of the lower respiratory tract, such as bronchoalveolar lavage fluid (BALF) or induced sputum (IS), are the most commonly utilized for the diagnosis of PcP. These specimens and others, such as open-lung biopsy (LB), transbronchial biopsy (TBB), and bronchial secretions (BS), are obtained by invasive techniques that, in addition to being onerous, are difficult to perform in patients with respiratory failure, children, and especially difficult to implement in low-to-middle income countries.
Lung biopsy (LB) is the gold standard procedure for the assessment of inflammatory lung conditions in immunocompromised patients [19], allowing the observation of the microorganism in more than 95% of the infection cases [20]. BALF, which is collected through fiber optic bronchoscopy, allows diagnosis in more than 80% of all patients with PcP, and in more than 95% of patients concurrently infected with HIV [21,22]. Sputum induction, which is obtained in a less invasive way through inhalation of 1.8% saline with the aid of an ultrasonic nebulizer, can be applied with good diagnostic yield in AIDS patients. However, in patients with other forms of immunodeficiency who generally have a lower burden of P. jirovecii, IS may have less diagnostic utility [23]. The non-specific staining of IS specimens detects Pneumocystis in 30-55% of cases of infection and the results are sometimes difficult to interpret [19,24]. Sensitivity can be improved to 60-97% with IS liquefaction using dithiothreitol, followed by cell sedimentation and analysis by immunofluorescence staining with anti-P. jirovecii monoclonal antibodies (MAb-IF) or PCR techniques [23,24,25]. As an alternative, other specimens like spontaneous sputum (SS), nasopharyngeal aspirate (NA), or oropharyngeal washing (OW), which are obtained less invasively, can also be used for diagnosis of PcP but have even lower diagnostic yields than BALF or IS [26]. Molecular techniques can be used as an alternative to improve the sensitivity of detection when using these less invasive respiratory specimens (SS, NA, OW) for PcP diagnosis [26,27,28,29,31], due to their lower fungal burden.
The diagnosis of extrapulmonary P. jirovecii infection is performed by the demonstration of developmental forms of the organism in the specific infected tissues. In these cases, non-specific as well as specific staining methods or a PCR method to detect Pneumocystis DNA can be applied, depending on the biological specimen available.
Staining methodologies. Several cytochemical staining methods exist for PcP diagnosis: Gomori methenamine silver (GMS); Gram-Weigert (GW); Giemsa; rapid Giemsa-like stains such as Diff-Quik (DQ); toluidine blue O (TBO); cresyl echt violet; and calcofluor white. These methods, despite being able to reveal the characteristic morphology of the cystic and/or the trophic forms of Pneumocystis, are non-specific and can stain other microorganisms.
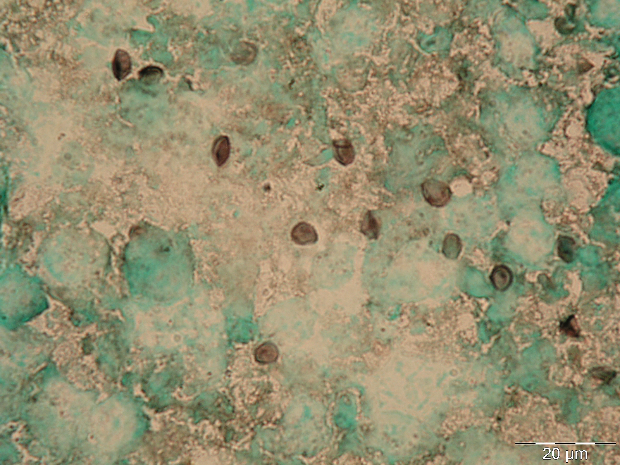
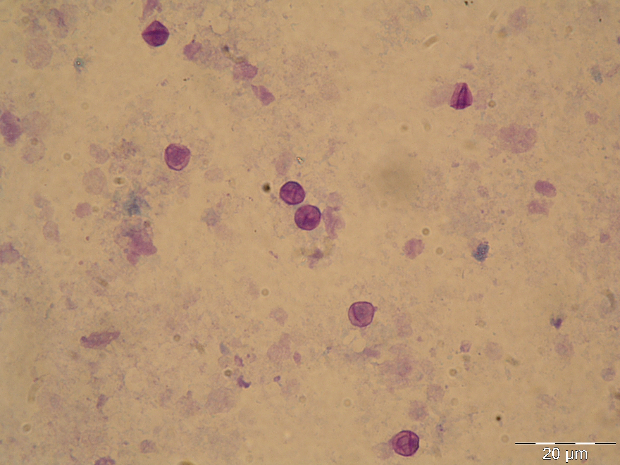
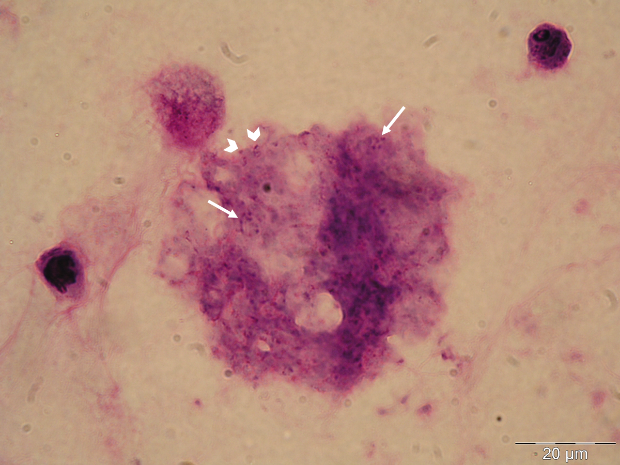
The GMS stain technique was first described by Gomori and then modified by Grocott (1955) and Musto (1982) [32,33] and was considered the gold standard for the diagnosis of PcP for many years. The reagent selectively stains the wall of the cystic forms of Pneumocystis, which appears dark brown (Figure 1A) [32]. Cystic forms of P. jirovecii can also be identified by GW and TBO staining, which have a good affinity for the cystic form wall components, selectively staining them in purple/blue or reddish violet (Figure 1B), respectively [34]. The cresyl echt violet technique (which is similar to TBO) and the chemifluorescent reagent calcofluor white (which binds non-specifically to β-linked polysaccharide polymers of the cell wall of Pneumocystis [35]) can also be used to identify Pneumocystis cystic forms, but not the other forms. Conversely, Giemsa and Diff-Quik do not stain sporocytic or cystic walls, but stain the nuclei of all Pneumocystis life cycle stages (Figure 1C) [36]. All of these methods can be applied in any kind of clinical specimen, but they are non-specific, presenting affinity to other lung pathogens. Therefore, the reading of the microscopic slides requires training and expertise, especially when dealing with patients with low fungal burden. When only non-specific staining methods are available, a good strategy for a more accurate PcP diagnosis is the concomitant use of a method that stains the nuclei of the developing forms (Giemsa or Diff-Quik) and another one that stains the wall of the cystic form (GMS, GW, TBO, calcofluor, or cresyl echt violet) in different smears from the same specimen [14,27].
|
(A)
|
(B)
|
|
(C)
|
(D)
|
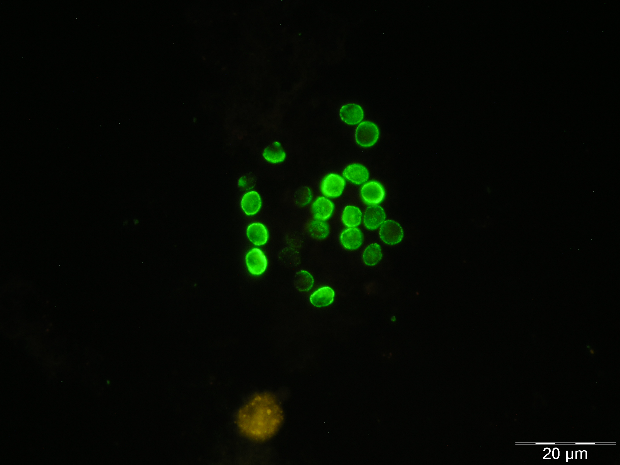
Figure 1 Rat and human respiratory specimens with Pneumocystis after proper staining (magnification X 1,000). (A) Rat-derived Pneumocystis cystic forms stained with Gomori methenamine silver (GMS). (B) Rat-derived Pneumocystis cystic forms stained with toluidine blue O (TBO). (C) Cluster of Pneumocystis jirovecii mature cystic forms (arrows) and trophic forms (arrowhead) in BALF stained with Giemsa. (D) Clustered cystic forms of Pneumocystis in BALF stained with IF-MAb anti-Pneumocystis jirovecii.
Specific staining techniques appeared with the development of monoclonal antibodies (MAbs) specific for P. jirovecii in 1986, which allowed the appearance of immunofluorescent techniques for the diagnosis of PcP [37,38]. These staining methods are considered more sensitive and specific than the cytochemical ones, and have begun to be applied as the preferred method in less than optimal specimens (IS, BS, NA, and OW) [24,25,38,39,40]. Currently, it is also possible to find some direct and indirect immunofluorescent assays (DFA and IFA, respectively) in the market for clinical use. The IFA identifies only the cystic forms while the DFA identifies both cystic and trophic forms. In these commercial kits, the anti-P. jirovecii MAbs are conjugated with fluorescein isothiocyanate (FITC) and, when they bind to the specific antigen of P. jirovecii, the cystic and/or trophic forms appear with a characteristic apple-green fluorescence when exposed to a given wavelength (Figure 1D). Nowadays, because of its reliability, these immunofluorescent stains are the most commonly used technique in the diagnosis of PcP [26,41,42,43].
Molecular biology methodologies. The ability to detect P. jirovecii DNA in clinical specimens by applying molecular tools has brought important advances in the diagnosis, epidemiology, and management of PcP in the last few decades. PCR methods allow the early detection of P. jirovecii DNA in respiratory specimens from patients that tested negative via a microscopic examination [29,44,45]. Thus, reducing the time from the onset of symptoms to the diagnosis and treatment of the disease, improving the prognosis, and avoiding the progression from the initial case of PcP to a serious illness with associated respiratory failure and significant mortality [12,26].
The efficiency of the molecular methods depends on the type of specimen analysed. Biopsies (which are highly invasive) are rarely used, while BALF is the standard specimen. Less invasive specimens like IS, NA, and OW can also be used but compromise the final overall sensitivity of the molecular PcP diagnostic protocol [26,44,46,47,48,49]. However, a positive PCR test associated with a negative microscopy causes ambiguity in the diagnosis of PcP, since it may reflect cases of either PcP or P. jirovecii colonization. This doubt can only be resolved by combining the laboratory results with the patient’s clinical picture [14]. In clinical practice, a case of P. jirovecii colonization is considered when P. jirovecii DNA is detected by PCR in a biological specimen of an immunocompromised or immunocompetent individual without clinical manifestations of PcP, and is also accompanied by a negative staining result [1,2,50]. In these cases, the false negative results of the microscopic examinations are due to the very low fungal burdens that are difficult to detect [51].
Different PCR techniques and gene targets have been reported for PcP molecular diagnosis [26,52,53,54,55,56,57,58,59,60,61,62,63,64,68,66,67,68,69]. The mitochondrial large subunit rRNA (mtLSUrRNA) nested-PCR procedure, with a detection threshold that can reach values of 0.5-1 organism/μL of sample [14,26,29], is the most used. Reports have shown that it is the most sensitive method among nine PCR assays evaluated for detection of P. jirovecii DNA [70]. This technique produced less false negative results and presented higher concordance with microscopic data than mtLSUrRNA single-PCR, internal transcribed spacers (ITS) nested-PCR, dihydropteroate synthase (DHPS) single- and nested-PCR, dihydrofolate reductase (DHFR) nested-PCR, major surface glycoprotein (MSG) heminested-PCR, 18S ribosomal RNA (18S rRNA) 1-tube nested-PCR, and 5S ribosomal RNA (5S rRNA) real-time quantitative PCR (RT-qPCR) [70]. In addition, the DHPS and DHFR PCR assays showed low diagnostic specificity in several studies [71,72,73]. The fact that mitochondrial mtLSU rRNA is a multicopy gene in the P. jirovecii genome, compared with other target genes that are single nuclear encoded genes (e.g. DHPS, DHFR, ITS), contributes largely to the higher successful amplification rates of the PCR procedures that target this gene, especially with the nested-PCR technique [26,56,74,75]. A bivariate meta-analysis and systematic review of 16 studies of PCR-based assays of a total of 1857 BALF from 1793 patients recorded a sensitivity of 98.3% (95% CI, 91.3%-99.7%) and specificity of 91.0% (95% CI, 82.7%-95.5%), which suggests that the application of these methodologies in BALF is a very accurate method for the diagnosis of PcP [47]. In particular, nested-PCR-based P. jirovecii detection techniques demonstrated a high sensitivity of 98% (95% CI, 76-100%) and a relatively medium specificity of 73% (95% CI, 53-86%) [47]. Once again, this could be explained by colonized/asymptomatic patients that test as false positive by nested-PCR although the symptomatic infection is not established. Therefore, this indicates that although nested-PCR protocols present high sensitivity in the detection of P. jirovecii DNA in respiratory specimens, a positive result with a negative microscopic examination always needs to be clinically investigated because it can correspond to a case of colonization, a symptomatic case, or even to a case of contamination [47,76].
P. jirovecii burden quantification is another theme of interest that prompted the development of several molecular strategies. The application of RT-qPCR protocols targeting the MSG multigene family, β-TUB, KEX1 genes, mtLSUrRNA, and cdc2 genes of P. jirovecii have been reported [45,74,77,78,79,80]. A meta-analysis study assessing the use of RT-qPCR protocols for the diagnosis of PcP in immunocompromised patients from 10 individual studies from 1990 to 2010 showed an overall sensitivity of 97% (95% CI, 93%-99%) and specificity of 94% (95% CI, 90%-96%) [31]. In the subgroup of HIV-infected patients, P. jirovecii DNA was detected with a sensitivity of 97% (95% CI, 93%-99%) and a specificity of 93% (95% CI, 89%-96%). The DNA detection in BALF demonstrated a sensitivity of 98% (95% CI, 94%-99%) and specificity of 93% (95% CI, 89%-96%) [31]. Despite good diagnostic accuracy results, RT-qPCR protocols still need to be investigated in new studies to identify any differences in the diagnostic performance of this method in HIV-infected versus other immunocompromised patients as well as in differentiating colonization from active disease [26]. Also, in order to improve accuracy in the management of PcP, thresholds should be assessed according to underlying diseases and other clinical and radiological parameters [80]. To guarantee clinical value of the results obtained, it is essential that both specificity and sensitivity are ≥95%. In addition, when interpreting the significance of the fungal burden, the quality of the biological specimen under study and the underlying condition should also be taken into consideration.
In conclusion, although nested-PCR and RT-qPCR assays (especially the ones targeting the mtLSUrRNA gene) are consistently indicated as the most sensitive and specific molecular tools for P. jirovecii DNA detection [46,51,57,73,77,81,82], it will be most helpful for diagnostic laboratories to choose standardized commercial tests that have developed a criterion for results interpretation.
2.2. Alternatives to the Classical Laboratory Diagnosis of PcP
The success of classical methods for PcP diagnosis depends on: resources and technology of the laboratory, the team’s experience, and the type of biological specimen to be analysed. The desire to use biological specimens obtained by less invasive techniques and less restricted technologies attracted the attention to and interest in the blood and serum. Blood specimens started to be tested as an alternative to respiratory specimens in the 1990s. At the beginning, once a bloodborne phase of the infection was suggested but never demonstrated, the attention was focused on the detection of P. jirovecii in blood by molecular methods such as PCR [87,88,89,90,91,92,93]. However, five in seven studies that applied this method showed low to very low sensitivity (0–30%), depending on the locus analysed [87,88,89,90,91,92,93]. Recently, other alternative strategies for a less-invasive PcP diagnosis emerged, based on the measurement of blood biomarkers that reflect the host–pathogen interaction [18,94,95]. Currently, the focus is on the detection of anti-P. jirovecii sera antibodies, because several reports using recombinant antigens of P. jirovecii and antibody immunodetection techniques have shown potential application in the diagnosis and epidemiological studies of PcP [96,97,98,99,100,101,102,103,104,105,106,107].
Blood biomarkers for non-invasive diagnosis of PcP. In the past few decades, many biomolecules that could be detected in the serum of patients were studied for use in the diagnosis of PcP. Molecules of the microorganism, such as (1-3)-β-d-glucan (BG), and also host molecules such as lactate dehydrogenase (LDH), Krebs von den Lungen-6 antigen (KL-6), as well as S-adenosylmethionine (SAM), have been considered as potential candidates for use as biomarkers in the serological diagnosis of PcP [18,94,95,108,109,110,111,112,113]. However, serum levels of all these metabolites are not strictly specific for P. jirovecii infection.
Although it is a structural molecule of the cell wall of P. jirovecii, the polysaccharide BG presents a panfungal character, which compromises the specificity rates for the diagnosis of PcP [94,112,114]. Additionally, it was suggested that false positive results can also be induced by the administration of certain agents that are filtered through cellulose membranes [115]. Otherwise, the fungal burden is another parameter that could compromise the serum concentrations of BG and, consequently, the reliability of the BG test for the diagnosis of PcP in patients without HIV infection, as the number of P. jirovecii organisms in the lungs of non-HIV-infected patients is usually lower than that observed in the lungs of HIV-infected patients [116,117]. Another major problem in the application of BG serum levels for PcP diagnosis is the absence of a definitive optimal cut-off limit, which would enable distinguishing between colonization and disease [18,118,119].
A variety of studies suggest that increased levels of the mucin-like glycoprotein KL-6 are present in patients with PcP, which is compatible with the fact that high serum levels of this glycoprotein are known to be an indicator of interstitial lung disease and acute lung damage [18,117]. However, since KL-6 is not a P. jirovecii-related molecule and is strongly expressed on type II alveolar pneumocytes and bronchiolar epithelial cells, increased levels may be more specific to underlying injury to the lung parenchyma rather than a specific marker of P. jirovecii infection. This could lead to false positive cases and consequently a lack of specificity to diagnose PcP [117].
The LDH enzyme is another biomarker that is widely expressed in human tissues, which is released into the blood stream after cell membrane damage. Therefore, although high serum levels of this biomarker are known to be elevated in patients with PcP, false positives can appear due to underlying lung injury and inflammation caused by P. jirovecii and/or other pathogen presence in the lungs, and even due to a variety of extrapulmonary disorders [18,120].
Finally, the applicability of SAM serum levels causes disagreement among several authors since conflicting data exist about the need for exogenous SAM and the presence of a functional SAM synthetase gene in P. jirovecii [110,116,121,122]. While some studies show SAM as an accurate biomarker for PcP [110,121], others show that this biomarker is unable to differentiate between patients with and without the disease [113,116], revealing very low diagnostic strength. Regardless, this biomarker is the one with the lowest potential to be used in the diagnosis of PcP, since the relationship between its serum levels and the presence of P. jirovecii infection is not well-defined.
Despite all of this, several studies have shown a good correlation between serum BG levels and disease severity, as well as demonstrating the ability to distinguish between P. jirovecii infection and other fungal infections, which is common in immunocompromised patients [18,113]. Two meta-analyses estimated that the measurement of BG serum levels for PcP diagnosis presents high sensitivity (95-96%), medium specificity (84-86%), and negative predictive value varying between 98.5% and 98.9% for a PcP prevalence of 20% in a population with a majority of HIV-infected patients [111,123]. However, recent studies reveal a lower sensitivity (85%) when testing non-HIV-infected patients [124]. As serum BG detection presents high sensitivity to diagnose PcP, a negative result allows exclusion of PcP in a patient at risk of the disease. However, the lack of specificity and the absence of a consensus threshold for its application in PcP diagnosis makes it impossible to consider a BG positive result by itself as diagnostic of PcP [112,125,126]. Even so, the serum measurement of this biomarker has shown to be able to contribute to PcP diagnosis as well as to exclude the disease when associated with PCR of upper respiratory tract specimens [126] and with other clinical diagnostic conditions indicative of PcP, especially in immunocompromised non-HIV-infected patients. Furthermore, the combination of BG and KL-6 tests showed great accuracy and can be an alternative for a minimally invasive and less costly diagnosis of PcP, compared to the classic diagnostic approach [112,113]. Table 2 briefly shows variables such as sensitivity, specificity, advantages, and disadvantages of the application of these blood biomarkers in the laboratory diagnosis of PcP [26,112,113].
Immunodiagnosis for less invasive detection of P. jirovecii infection. Reports of PcP in patients with genetic mutations affecting immunoglobulin production [127], along with reports of high incidence of positive serology for P. jirovecii in healthy adults [128], highlights the important role of humoral immunity during PcP and supports the idea that a serological test is viable.
Currently, several studies have shown that human serum antibodies are able to recognize recombinant antigens of P. jirovecii proteins [96,97,98,99,100,101,102,103,104,105,106,107]. Until now, the major surface glycoprotein (MSG) has received the most attention. This is a protein highly specific to this pathogen that plays a central role in the interaction of Pneumocystis with its host, eliciting humoral and cellular protective immune responses [107,129]. The three individual recombinant fragments that cover the entire length of MSG (MsgA, MsgB, and MsgC) were analysed in human serum specimens for P. jirovecii antibody detection and this analysis revealed that the carboxyl-terminal domain (MsgC) is the most conserved and reactive region of this glycoprotein [96,98,103,104,105]. This data suggests that epitopes that stimulate at least part of the human antibodies against P. jirovecii may be located in this region. Taking this into consideration, a recombinant synthetic antigen with three antigenic regions of the Msg protein, specifically, one from the terminal portion of MsgB and two from MsgC (Figure 2), was recently designed and produced [107]. The antigen was purified and applied as an antigenic tool in an ELISA assay (Figure 3) for anti-P. jirovecii antibody detection. The results showed increased IgM anti-P. jirovecii levels in PcP patients compared with patients without PcP. This ELISA assay presented a sensitivity of 100% and a specificity of 80.8% when associated with the clinical diagnosis of PcP of each patient [107]. These results suggest that, based on the immunogenic behaviour of P. jirovecii proteins, newly recombinant synthetic multi-epitope antigenic peptides (RSAs) are one of the most promising approaches to developing diagnostic platforms for routine screening of PcP [130,131,132]. This technology is based on synthetic amino acid sequences designed to contain more than one reactive region of each selected antigen, increasing the sensitivity and specificity of the serological test as well as making it cheaper and easier to standardize [130,131].
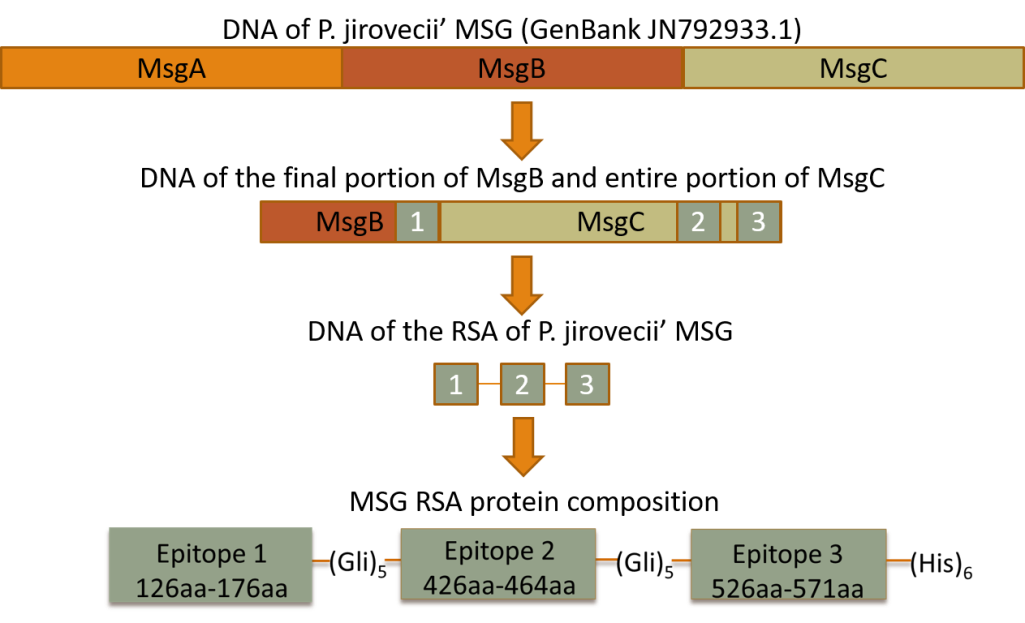
Figure 2 Scheme illustrating the design and composition of the three selected epitopes of the MSG RSA applied in an ELISA platform for PcP diagnosis.
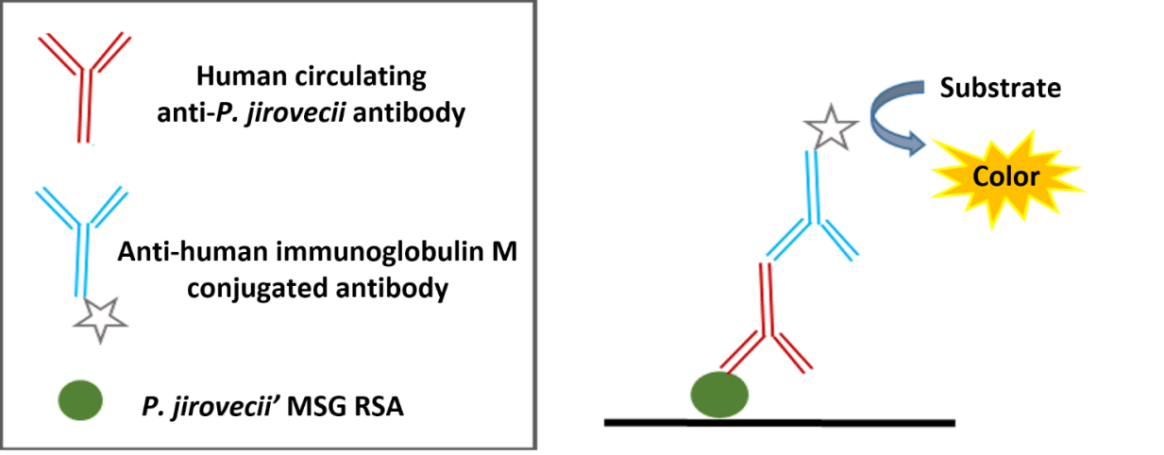
Figure 3 Indirect ELISA assay developed for serological diagnosis of PcP.
More recently, the attention of researchers began to focus on other proteins of P. jirovecii such as kexin-like serine protease (KEX1) and the GPI-anchored cell surface protein MEU10. KEX1 is a nuclear single-copy gene involved in the processing of proteins that maintain P. jirovecii cell surface integrity such as the proteolytic processing of MSG [104,133]. Recombinant KEX1 segments have been developed to study humoral responses to P. jirovecii and some variants were found useful in identifying acute cases of PcP, or even as indicators of subsequent risk of PcP [98,103,104,105]. Also, a new MEU10 antigenic epitope, a GPI-anchored protein that appears to be in the surface of both the trophic and the cystic forms and is also conserved in P. jirovecii, was recently described to be capable of inducing a humoral response during Pneumocystis infection [134]. Thus, there is a need to better characterize the serologic responses to epitopes from these proteins and use standardized antigen preparations to assess the host immune response to P. jirovecii infection.
Moreover, a new tool based on innovative nanotechnology approaches is being developed. Biosensors become faster, more sensitive, and flexible when gold nanoparticles (AuNPs) are used as tags or labels [135,136,137,138], because AuNPs have high surface areas and unique physicochemical properties, e.g., tunable bright color, that make them ideal candidates for developing biomarker platforms [137]. AuNPs can be used in the development of methods suitable for clinical diagnosis, where AuNPs serve as signal transducers and as scaffolds for bio-recognition [138]. Thus, the possibility of combining the powerful technologies of recombinant synthetic multi-epitope antigens (RSAs) with the extraordinary properties and high sensitivity of AuNPs may have a major impact on the point-of-care diagnosis of PcP. The innovative nature of this approach is the use of RSAs in association with AuNPs to design new platforms for PcP diagnosis at point-of-care to detect specific anti-P. jirovecii antibodies using a simple, fast, sensitive, specific, and inexpensive solid-phase (e.g., strip-based) test and a readily available, less expensive, and minimally invasive sample, such as blood. Preliminary results [139] not yet published have shown that this approach is possible to achieve, and that it could result in a point-of-care platform that will enable a faster and cheaper screening and diagnosis of this opportunistic infectious disease, helping to refine therapeutic interventions, improve disease control, and provide retrenchment of healthcare systems.
Therefore, it is expected that in the near future this and other approaches will emerge, based on these or other measurable serum biomarkers in the search for new tools for definitive, non-invasive PcP diagnosis.
Table 2 Characteristics of the tests based on measurement of blood biomarkers for PcP diagnosis (adapted from [26]).

3. Conclusions
Despite the availability of cART and prophylaxis against PcP, this disease remains a significant cause of mortality and morbidity in HIV-infected and non-HIV-infected patients.
Early diagnosis of PcP is crucial for a timely implementation of treatment and for a better prognosis. Lately, new approaches for the diagnosis of PcP emerged. Several clinical support tools, such as clinical history, physical examination, and nonspecific radiological and laboratory tests may suggest the disease, but none of them is decisive. The definitive diagnosis of PcP still depends on specific laboratory methodologies, which have improved dramatically in the last 30 years. The classical methods, based on cytochemical or immunofluorescent staining and molecular biology assays, could be applied to a variety of more or less invasive respiratory specimens, which influence the sensitivity, specificity, cost, and complexity of the diagnosis. New alternatives based on the measurement of blood biomarkers of infection, such as the detection of anti-P.jirovecii antibodies, continue to be investigated. When selecting which laboratorial method to utilize, the local incidence of PcP, the type of biological specimen available, as well as the level of local resources and expertise should be taken into consideration. Improving the accuracy of P. jirovecii detection, enabling PcP diagnosis through less invasive biological specimens, and developing a way to discriminate between patients with PcP and those who are carriers still need to be further researched. New diagnostic platforms based upon nanotechnology using non-invasive biological specimens are promising tools for an easier and cheaper detection of PcP biomarkers, enabling an early implementation of therapeutic and prophylactic measures and facilitating disease control, especially in resource-limited settings. However, these innovative approaches still need optimization and validation for their application in PcP diagnosis and implementation in clinical practice.
Acknowledgments
Authors thank Moussa Elbayoumy, MD, for critical review and linguistic editing.
Author Contributions
Ana Luísa Tomás and Olga Matos contributed equally to this work.
Competing Interests
The authors have declared that no competing interests exist.
References
- Barry SM, Johnson MA. Pneumocystis carinii pneumonia: a review of current issues in diagnosis and management. HIV Med. 2001; 2: 123-132. [CrossRef] [Google scholar] [PubMed]
- Morris A, Lundgren JD, Masur H, Walzer PD, Hanson DL, Frederick T, et al. Current epidemiology of Pneumocystis pneumonia. Emerg Infect Dis. 2004; 10: 1713-1720. [CrossRef] [Google scholar] [PubMed]
- Matos O, Costa MC, Correia I, Monteiro P, Vieira JR, Soares J, et al. Pneumocystis jiroveci infection in immunocompetent patients with pulmonary disorders, in Portugal. Acta Med Port. 2006; 19: 121-126. [Google scholar]
- Raviglione MC. Extrapulmonary PcP: the first 50 cases. Rev Infect Dis. 1990; 12: 1127-1138. [CrossRef] [Google scholar] [PubMed]
- European Centre for Disease Prevention and Control/WHO Regional Office for Europe. HIV/AIDS surveillance in Europe 2017 – 2016 data. Stockholm: ECDC; 2017. [Google scholar]
- French N, Kaleebu P, Pisani E, Whitworth JAG. Human immunodeficiency virus (HIV) in developing countries. Ann Trop Med Parasit. 2006; 100: 433-454. [CrossRef] [Google scholar] [PubMed]
- Matos O, Esteves F. Epidemiology and clinical relevance of Pneumocystis jirovecii Frenkel, 1976 dihydropteroate synthase gene mutations. Parasite. 2010; 17: 219-232. [CrossRef] [Google scholar] [PubMed]
- de Armas Rodríguez Y, Wissmann G, Müller AL, Pederiva MA, Brum MC, Brackmann RL, et al. Pneumocystis jirovecii pneumonia in developing countries. Parasite. 2011; 18: 219-228. [CrossRef] [Google scholar] [PubMed]
- Matos O. Pneumocystis jirovecii pneumonia in Africa: impact and implications of highly sensitive diagnostic technologies. N Am J Med Sci. 2012; 4: 486-487. [Google scholar]
- Hughes WT. Pneumocystis pneumonitis in non-HIV-infected patients: update. In: Pneumocystis Pneumonia, 3rd edn. New York: Marcel Dekker; 2005. p. 407-434. [Google scholar]
- Mocroft A, Katlama C, Johnson AM, Pradier C, Antunes F, Mulcahy F, et al. AIDS across Europe, 1994-98: the EuroSIDA study. Lancet. 2000; 356:291-296. [CrossRef] [Google scholar] [PubMed]
- Huang L. Clinical Presentation and Diagnosis of Pneumocystis Pneumonia in HIV-Infected Patients. In: Pneumocystis Pneumonia, 3rd edn. New York: Marcel Dekker; 2005. p. 349-406. [Google scholar]
- Walzer PD, Evans HE, Copas AJ, Edwards SG, Grant AD, Miller RF. Early predictors of mortality from Pneumocystis jirovecii pneumonia in HIV-infected patients: 1985-2006. Clin Infect Dis. 2008; 46: 625-633. [CrossRef] [Google scholar] [PubMed]
- Matos O, Tomás AL, Antunes F. Pneumocystis jirovecii and PcP. In: Current Progress in Medical Mycology. Cham: Springer International Publishing; 2017. p. 215-254. [CrossRef] [Google scholar]
- Aliouat-Denis CM, Chabe M, Demanche C, Aliouat el M, Viscogliosi E, Guillot J, et al. Pneumocystis species, co-evolution and pathogenic power. Infect Genet Evol. 2008; 8: 708-726. [CrossRef] [Google scholar] [PubMed]
- Schildgen V, Mai S, Khalfaoui S, Lüsebrink J, Pieper M, Tillmann RL, et al. Pneumocystis jirovecii can be productively cultured in differentiated CuFi-8 airway cells. MBio. 2014; 5: e01186-14. [CrossRef] [Google scholar] [PubMed]
- Liu Y, Fahle GA, Kovacs JA. Inability to Culture Pneumocystis jirovecii. MBio. 2018; 9: e00939-18. [CrossRef] [Google scholar] [PubMed]
- Tasaka S, Hasegawa N, Kobayashi S, Yamada W, Nishimura T, Takeuchi T et al. Serum indicators for the diagnosis of Pneumocystis pneumonia. Chest. 2007; 131: 1173-1180. [CrossRef] [Google scholar] [PubMed]
- Rodriguez M, Fishman JA. Prevention of infection due to Pneumocystis spp. in human immunodeficiency virus-negative immunocompromised patients. Clin Microbiol Rev. 2004; 17: 770-782. [CrossRef] [Google scholar] [PubMed]
- Rossiter SJ, Miller DC, Churg AM et al. Open lung biopsy in the immunosuppressed patient. Is it really beneficial? J Thorac Cardiovasc Surg. 1979; 77: 338-345. [Google scholar]
- Baughman RP, Dohn MN, Frame PT. The continuing utility of bronchoalveolar lavage to diagnose opportunistic infection in AIDS patients. Am J Med. 1994; 97: 515-522. [CrossRef] [Google scholar] [PubMed]
- Fishman JA. Pneumocystis carinii and parasitic infection in the immunocompromised host. In: Clinical approach to infection in the compromised host. New York: Kluwer Academic/Plenum Publishers; 2002. p. 265-334. [CrossRef] [Google scholar]
- LaRocque RC, Katz JT, Perruzzi P, et al. The utility of sputum induction for diagnosis of Pneumocystis pneumonia in immunocompromised patients without human immunodeficiency virus. Clin Infect Dis. 2003; 37: 1380-1383. [CrossRef] [Google scholar] [PubMed]
- Kovacs JA, Ng VL, Masur H, et al. Diagnosis of Pneumocystis carinii pneumonia: improved detection in sputum with use of monoclonal antibodies. New Engl J Med. 1988; 318: 589-593. [Google scholar]
- Ng VL, Virani NA, Chaisson RE, et al. Rapid detection of Pneumocystis carinii using a direct fluorescent monoclonal antibody stain. J Clin Microbiol. 1990; 28: 2228-2233. [Google scholar]
- Matos O, Esteves F. Laboratory diagnosis of Pneumocystis jirovecii pneumonia. In: Microbiology of respiratory system infection. United Kingdom: Elsevier 2016. p. 185-210. [CrossRef] [Google scholar]
- Wakefield AE, Miller RF, Guiver LA, Hopkin JM. Oropharyngeal samples for detection of Pneumocystis carinii by DNA amplification. Int J Med. 1993; 86: 401-406. [Google scholar]
- Respaldiza N, Montes-Cano MA, Friaza V, Muñoz-Lobato F, Medrano FJ, Varela JM, et al. Usefulness of oropharyngeal washings for identifying Pneumocystis jirovecii carriers. J Eukaryot Microbiol. 2006; 53: 100-101. [CrossRef] [Google scholar] [PubMed]
- Durand-Joly I, Chabé M, Soula F, Delhaes L, Camus D, Dei-Cas E. Molecular diagnosis of Pneumocystis pneumonia (PcP). FEMS Immunol Med Microbiol. 2005; 45: 405–410. [CrossRef] [Google scholar] [PubMed]
- Larsen HH, Huang L, Kovacs JA, Crothers K, Silcott VA, Morris A et al. A prospective, blinded study of quantitative touch-down polymerase chain reaction using oral-wash samples for diagnosis of Pneumocystis pneumonia in HIV-infected patients. J Infect Dis. 2004; 189: 1679–1683. [CrossRef] [Google scholar] [PubMed]
- Summah H, Zhu YG, Falagas ME, Vouloumanou EK, Qu JM. Use of real-time polymerase chain reaction for the diagnosis of Pneumocystis pneumonia in immunocompromised patients: a meta-analysis. Chin Med J (Engl). 2013; 126: 1965-1973. [Google scholar]
- Grocott RG. A stain for fungi in tissue sections and smears using Gomori’s methenamine-silver nitrate technic. Am J Clin Pathol. 1955; 25: 975-979. [CrossRef] [Google scholar] [PubMed]
- Musto L, Flanigan M, Elbadawi A. Ten-minute silver stain for Pneumocystis carinii and fungi in tissue sections. Arch Pathol Lab Med. 1982; 106: 292-294. [Google scholar]
- Gosey LL, Howard RM, Witebsky FG, Ognibene FP, Wu TC, Gill VJ, et al. Advantages of a modified toluidine blue O stain and bronchoalveolar lavage for the diagnosis of Pneumocystis carinii pneumonia. J Clin Microbiol. 1985; 22: 803-807. [Google scholar]
- Baselski VS, Robison MK, Pifer LW, Woods DR. Rapid detection of Pneumocystis carinii in bronchoalveolar lavage samples by using Cellufluor staining. J Clin Microbiol. 1990; 28: 393-394. [Google scholar]
- Holten-Andersen W, Kolmos HJ. Comparison of methenamine silver nitrate and Giemsa stain for detection of Pneumocystis carinii in bronchoalveolar lavage specimens from HIV infected patients. APMIS. 1989; 97: 745-747. [CrossRef] [Google scholar] [PubMed]
- Kovacs JA, Gill V, Swan JC, Ognibene F, Shelhamer J, Parrillo JE, et al. Prospective evaluation of a monoclonal antibody in diagnosis of Pneumocystis carinii pneumonia. Lancet. 1986; 2: 1-3. [CrossRef] [Google scholar] [PubMed]
- Kovacs JA, Gill VJ, Meshnick S, Masur H. New insights into transmission, diagnosis, and drug treatment of Pneumocystis carinii pneumonia. J Am Med Assoc. 2001; 286: 2450–2460. [CrossRef] [Google scholar] [PubMed]
- Ng VL, Yajko DM, Mcphaul LW, Gartner I, Byford B, Goodman CD, et al. Evaluation of an indirect fluorescentantibody stain for detection of Pneumocystis carinii in respiratory specimens. J Clin Microbiol. 1990; 28: 975-979. [Google scholar]
- Cregan P, Yamamoto A, Lum A, VanDerHeide T, MacDonald M, Pulliam L. Comparison of four methods for rapid detection of Pneumocystis carinii in respiratory specimens. J Clin Microbiol. 1990; 28: 2432-2436. [Google scholar]
- Baughman RP, Strohoper SS, Clinton BA, Nickol AD, Frame PI. The use of an indirect fluorescent antibody test for detecting Pneumocystis carinii. Arch Pathol Lab Med. 1989; 113: 1062-1065. [Google scholar]
- Lautenschlager I, Lyytikainen O, Jokipii L, Jokipii A, Maiche A, Ruutu T, et al. Immunodetection of Pneumocystis carinii in bronchoalveolar lavage specimens compared with methenamine silver stain. J Clin Microbiol. 1996; 34: 728-730. [Google scholar]
- Bava AJ, Cattaneo S, Bellegarde E. Diagnosis of pulmonary PcP by microscopy on wet mount preparations. Rev Inst Med Trop S Paulo. 2002; 44: 279-282. [CrossRef] [Google scholar] [PubMed]
- Matos O, Costa MC, Lundgren B, Caldeira L, Aguiar P, Antunes F. Effect of oral washes on the diagnosis of Pneumocystis carinii pneumonia with a low parasite burden and on detection of organisms in subclinical infections. Eur J Clin Microbiol Inf Dis. 2001; 20: 573-575. [CrossRef] [Google scholar] [PubMed]
- Rohner P, Jacomo V, Studer R, Schrenzel J, Graf JD. Detection of Pneumocystis jirovecii by two staining methods and two quantitative PCR assays. Infection. 2009; 37: 261-265. [CrossRef] [Google scholar] [PubMed]
- Durand-Joly I, Chabé M, Soula F, Delhaes L, Camus D, Dei-Cas E. Molecular diagnosis of Pneumocystis pneumonia (PcP). FEMS Immunol Med Microbiol. 2005; 45: 405–410. [CrossRef] [Google scholar] [PubMed]
- Fan LC, Lu HW, Cheng KB, Li HP, Xu JF. Evaluation of PCR in bronchoalveolar lavage fluid for diagnosis of Pneumocystis jirovecii pneumonia: a bivariate meta-analysis and systematic review. Plos One. 2013; 8: e73099. [CrossRef] [Google scholar] [PubMed]
- Durand-Joly I, Soula F, Chabé M, Dalle JH, Lafitte JJ, Senechal M. Longterm colonization with Pneumocystis jirovecii in hospital staffs: a challenge to prevent nosocomial PcP. J Eukaryot Microbiol. 2003; 50: 614-615. [CrossRef] [Google scholar] [PubMed]
- Arcenas RC, Uhl JR, Buckwalter SP, Limper AH, Crino D, Roberts GD. A real-time polymerase chain reaction assay for detection of Pneumocystis from bronchoalveolar lavage fluid. Diagn Microbiol Infect Dis. 2006; 54: 169-175. [CrossRef] [Google scholar] [PubMed]
- Morris A, Sciurba FC, Norris KA. Pneumocystis: a novel pathogen in chronic obstructive pulmonary disease?. COPD. 2008; 5: 43-51a. [CrossRef] [Google scholar] [PubMed]
- Vidal S, de la Horra C, Martín J, Montes-Cano MA, Rodríguez E, Respaldiza N. Pneumocystis jirovecii colonisation in patients with interstitial lung disease. Clin Microbiol Infect. 2006; 12: 231-235. [CrossRef] [Google scholar] [PubMed]
- Mei Q, Gurunathan S, Masur H, Kovacs JA. Failure of co-trimoxazole in Pneumocystis carinii infection and mutations in dihydropteroate synthase gene. Lancet. 1998; 351: 1631-1632. [CrossRef] [Google scholar] [PubMed]
- Walker DJ, Wakefield AE, Dohn MN, Miller RF, Baughman RP, Hossler PA. Sequence polymorphisms in the Pneumocystis carinii cytochrome b gene and their association with atovaquone prophylaxis failure. J Infect Dis. 1998; 178: 1767-1775. [CrossRef] [Google scholar] [PubMed]
- Ma L, Borio L, Masur H, Kovacs JA. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J Infect Dis. 1999; 180: 1969-1978. [CrossRef] [Google scholar] [PubMed]
- Kutty G, Kovacs JA. A single-copy gene encodes Kex1, a serine endoprotease of Pneumocystis jiroveci. Infect Immun. 2003; 71: 571-574. [CrossRef] [Google scholar] [PubMed]
- Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Miller RF, Moxon ER, et al. Amplification of mitochondrial ribosomal RNA sequences from Pneumocystis carinii DNA of rat and human origin. Mol Biochem Parasitol. 1990; 43: 69-76. [CrossRef] [Google scholar] [PubMed]
- Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Miller RF, Moxon ER, et al. Detection of Pneumocystis carinii with DNA amplification. Lancet. 1990; 336: 451-453. [CrossRef] [Google scholar] [PubMed]
- Kaiser K, Rabodonirina M, Mayencon M, Picot S. Evidence for cdc2 gene in Pneumocystis carinii hominis and its implication for culture. AIDS. 1999; 13: 419-420. [CrossRef] [Google scholar] [PubMed]
- Wada M, Nakamura Y. Unique telomeric expression site of major-surface-glycoprotein genes of Pneumocystis carinii. DNA Res. 1996; 3: 55-64. [CrossRef] [Google scholar] [PubMed]
- Hunter JA, Wakefield AE. Genetic divergence at the mitochondrial small subunit ribosomal RNA gene among isolates of Pneumocystis carinii from five mammalian host species. J Eukaryot Microbiol. 1996; 43: 24S-245. [CrossRef] [Google scholar] [PubMed]
- Denis CM, Mazars E, Guyot K, Odberg-Ferragut C, Viscogliosi E, Dei-CAS E, et al. Genetic divergence at the SODA locus of six different formae speciales of Pneumocystis carinii. Med Mycol. 2000; 38: 289-300. [CrossRef] [Google scholar] [PubMed]
- Stringer JR, Stringer SL, Zhang J, Baughman R, Smulian AG, Cushion MT. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993; 40: 733-741. [CrossRef] [Google scholar] [PubMed]
- Edlind TD, Bartlett MS, Weinberg GA, Prah GN, Smith JW. The beta-tubulin gene from rat and human isolates of Pneumocystis carinii. Mol Microbiol. 1992; 6: 3365-3373. [CrossRef] [Google scholar] [PubMed]
- Kutty G, Huang SN, Kovacs JA. Characterization of thioredoxin reductase genes (trr1) from Pneumocystis carinii and Pneumocystis jiroveci. Gene. 2003; 310: 175-183. [CrossRef] [Google scholar] [PubMed]
- Banerji S, Lugli EB, Miller RF, Wakefield AR. Analysis of genetic diversity at the arom locus in isolates of Pneumocystis carinii. J Eukaryot Microbiol. 1995; 42: 675-679. [CrossRef] [Google scholar] [PubMed]
- Liu Y, Rocourt M, Pan S, Liu C, Leibowitz MJ. Sequence and variability of the 5.8S and 26S rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 1992; 20: 3763-3772. [CrossRef] [Google scholar] [PubMed]
- Lu JJ, Bartlett MS, Shaw MM, Queener SF, Smith JW, Ortiz-Rivera M, et al. Typing of Pneumocystis carinii strains that infect humans based on nucleotide sequence variations of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1994; 32: 2904-2912. [Google scholar]
- Mazars E, Odberg-Ferragut C, Dei-Cas E, Fourmaux MN, Aliouat EM, Brun-Pascaud M, et al. Polymorphism of the thymidylate synthase gene of Pneumocystis carinii from different host species. J Eukaryot Microbiol. 1995; 42: 26-32. [CrossRef] [Google scholar] [PubMed]
- Garbe TR, Stringer JR. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect Immun. 1994; 62: 3092-3101. [Google scholar]
- Totet A, Pautard JC, Raccurt C, Roux P, Nevez G. Genotypes at the internal transcribed spacers of the nuclear rRNA operon of Pneumocystis jiroveci in nonimmunosuppressed infants without severe pneumonia. J Clin Microbiol. 2003; 41: 1173-1180. [CrossRef] [Google scholar] [PubMed]
- Cissé OH, Pagni M, Hauser PM. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. MBio. 2012. 4: e00428-12. [CrossRef] [Google scholar] [PubMed]
- Lu JJ, Chen CH, Bartlett MS, Smith Jw, Lee CH. Comparison of six different PCR methods for detection of Pneumocystis carinii. J Clin Microbiol. 1995; 33: 2785-2788. [Google scholar]
- Robberts FJ, Liebowitz LD, Chalkley LJ. Polymerase chain reaction detection of Pneumocystis jiroveci: evaluation of 9 assays. Diagn Microbiol Infect Dis. 2007; 58:385-92. [CrossRef] [Google scholar] [PubMed]
- Esteves F, Gaspar J, de Sousa B, Antunes F, Mansinho K, Matos O. Pneumocystis jirovecii multilocus genotyping in pooled DNA samples: a new approach for clinical and epidemiological studies. Clin Microbiol Infect. 2012; 18: E177-E184. [CrossRef] [Google scholar] [PubMed]
- Olsson M, Strålin K, Holmberg H. Clinical significance of nested polymerase chain reaction and immunofluorescence for detection of Pneumocystis carinii pneumonia. Clin Microbiol Infect. 2001; 7: 492-497. [CrossRef] [Google scholar] [PubMed]
- Moonens F, Liesnard C, Brancart F, Van Vooren JP, Serruys F. Rapid simple and nested polymerase chain reaction for the diagnosis of Pneumocystis carinii pneumonia. Scand J Infect Dis. 1995; 27: 358-362. [CrossRef] [Google scholar] [PubMed]
- Botterel F, Cabaret O, Foulet F, Cordonnier C, Costa JM, Bretagne S. Clinical significance of quantifying Pneumocystis jirovecii DNA by using real-time PCR in bronchoalveolar lavage fluid from immunocompromised patients. J Clin Microbiol. 2012; 50: 227-231. [CrossRef] [Google scholar] [PubMed]
- McTaggart LR, Wengenack NL, Richardson SE. Validation of the mycassay Pneumocystis kit for detection of Pneumocystis jirovecii in bronchoalveolar lavage specimens by comparison to a laboratory standard of direct immunofluorescence microscopy, real-time PCR, or conventional PCR. J Clin Microbiol. 2012; 50: 1856-1859. [CrossRef] [Google scholar] [PubMed]
- Larsen HH, Masur H, Kovacs JA, Gill VJ, Silcott VA, Kogulan P, et al. Development and evaluation of a quantitative, touchdown, real-time PCR assay for diagnosing Pneumocystis carinii pneumonia. J Clin Microbiol. 2002; 40: 490-494. [CrossRef] [Google scholar] [PubMed]
- Montesinos I, Brancart F, Schepers K, Jacobs F, Denis O, Delforge ML. Comparison of 2 real-time PCR assays for diagnosis of Pneumocystis jirovecii pneumonia in human immunodeficiency virus (HIV) and non-HIV immunocompromised patients. Diagn Microbiol Infect Dis. 2015; 82: 143-147. [CrossRef] [Google scholar] [PubMed]
- Wakefield AE. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996; 34: 1754-1759. [Google scholar]
- Lu JJ, Lee CH. Pneumocystis pneumonia. J Formos Med Assoc. 2008; 107: 830-842. [CrossRef] [Google scholar] [PubMed]
- Procop GW, Haddad S, Quinn J, Wilson ML, Henshaw NG, Reller LB, et al. Detection of Pneumocystis jiroveci in respiratory specimens by four staining methods. J Clin Microbiol. 2004; 42: 3333-3335. [CrossRef] [Google scholar] [PubMed]
- Aderaye G, Woldeamanuel Y, Asrat D, Lebbad M, Beser J, Worku A, et al. Evaluation of Toluidine Blue O staining for the diagnosis of Pneumocystis jiroveci in expectorated sputum sample and bronchoalveolarlavage from HIV-infected patients in a tertiary care referral center in Ethiopia. Infection. 2008; 36: 237-243. [CrossRef] [Google scholar] [PubMed]
- Raab SS, Cheville JC, Bottles K, Cohen MB. Utility of Gomori methenamine silver stains in bronchoalveolar lavage specimens. Mod Pathol. 1994; 7: 599-604. [Google scholar]
- Galan F, Oliver JL, Roux P, Poirot JL, Bereziat G. Detection of Pneumocystis carinii DNA by polymerase chain reaction compared to direct microscopy and immunofluorescence. J Protozool. 1991; 38: 199S-200S. [Google scholar]
- Schluger N, Sepkowitz K, Armstrong D, Bernard E, Rifkin M, Cerami A, et al. Detection of Pneumocystis carinii in serum of AIDS patients with Pneumocystis pneumonia by the polymerase chain reaction. J Protozool. 1991; 38: 240S-242S. [Google scholar]
- Lipschik GY, Gill VJ, Lundgren JD, Andrawis VA, Nelson NA, Nielsen JO, et al. Improved diagnosis of Pneumocystis carinii infection by polymerase chain reaction on induced sputum and blood. Lancet. 1992; 340: 203-206. [CrossRef] [Google scholar] [PubMed]
- Roux P, Lavrard I, Poirot JL, Chouaid C, Denis M, Olivier JL, et al. Usefulness of PCR for detection of Pneumocystis carinii DNA. J Clin Microbiol. 1994; 32: 2324-2326. [Google scholar]
- Atzori C, Lu JJ, Jiang B, Martlett MS, Orlando G, Queener SF, et al. Diagnosis of Pneumocystis carinii pneumonia in AIDS patients by using polymerase chain reactions on serum specimens. J Infect Dis. 1995; 172: 1623-1626. [CrossRef] [Google scholar] [PubMed]
- Tamburrini E, Mencarini P, Visconti E, Zolfo M, De Luca A, Siracusano A, et al. Detection of Pneumocystis carinii DNA in blood by PCR is not of value for diagnosis of P. carinii pneumonia. J Clin Microbiol. 1996; 34: 1586-1588. [Google scholar]
- Matos O, Lundgren B, Caldeira L, Mansinho K, Aguiar P, Forte M, et al. Evaluation of a nested PCR for detection of Pneumocystis carinii in serum from imunocompromised patients. J Euk Microbiol. 1999; 46: 104S-105S. [Google scholar]
- Rabodonirina M, Cotte L, Boibieux A, Kaiser K, Mayencon M, Raffenot D, et al. Detection of Pneumocystis carinii DNA in blood specimens from human immunodeficiency virus-infected patients by nested PCR. J Clin Microbiol. 1999; 37: 127-131. [Google scholar]
- Finkelman MA. Pneumocystis jirovecii infection: Cell wall (1-3)-D-glucan biology and diagnostic utility. Crit Rev Microbiol. 2010; 36: 271-281. [CrossRef] [Google scholar] [PubMed]
- Morris AM, Masur H. A serologic test to diagnose Pneumocystis pneumonia: are we there yet? Clin Infect Dis. 2011; 53: 203-204. [CrossRef] [Google scholar] [PubMed]
- Daly KR, Fichtenbaum CJ, Tanaka R, Linke MJ, O’Bert R, Thullen TD. Serologic responses to epitopes of the major surface glycoprotein of Pneumocystis jirovecii differ in human immunodeficiency virus-infected and uninfected persons. J Infect Dis. 2002; 186: 644-651 [CrossRef] [Google scholar] [PubMed]
- Bishop LR, Kovacs JA. Quantitation of anti-Pneumocystis jirovecii antibodies in healthy persons and immunocompromised patients. J Infect Dis. 2003; 187: 1844-1848. [CrossRef] [Google scholar] [PubMed]
- Daly KR, Koch J, Levin L, Walzer PD. Enzyme-linked immunosorbent assay and serologic responses to Pneumocystis jirovecii. Emerg Infect Dis. 2004; 10: 848-854. [CrossRef] [Google scholar] [PubMed]
- Daly KR, Koch JV, Shire NJ, Levin L, Walzer PD. Human immunodeficiency virus-infected patients with prior Pneumocystis pneumonia exhibit increased serologic reactivity to several major surface glycoprotein clones. Clin Vaccine Immunol. 2006; 13: 1071-1078. [CrossRef] [Google scholar] [PubMed]
- Daly KR, Koch JV, Respaldiza N, de la Horra C, Montes-Cano MA, Medrano FJ. Geographical variation in serological responses to recombinant Pneumocystis jirovecii major surface glycoprotein antigens. Clin Microbiol Infect. 2009; 15: 937-942. [CrossRef] [Google scholar] [PubMed]
- Tipirneni R, Daly KR, Jarlsberg LG, Koch JV, Swartzman A, Roth BM. Healthcare worker occupation and immune response to Pneumocystis jirovecii. Emerg Infect Dis. 2009; 15: 1590-1597. [CrossRef] [Google scholar] [PubMed]
- Walzer PD, Djawe K, Levin L, Daly KR, Koch J, Kingsley L, et al. Long-term serologic responses to the Pneumocystis jirovecii major surface glycoprotein in HIV-positive individuals with and without P. jirovecii infection. J Infect Dis. 2009; 199: 1335-1344. [CrossRef] [Google scholar] [PubMed]
- Djawe K, Huang L, Daly KR, Levin L, Koch J, Schwartzman A, et al. Serum antibody levels to the Pneumocystis jirovecii major surface glycoprotein in the diagnosis of P. jirovecii pneumonia in HIV+ patients. Plos One. 2010; 5: e14259. [CrossRef] [Google scholar] [PubMed]
- Gingo MR, Lucht L, Daly KR, Djawe K, Palella FJ, Abraham AG, et al. Serologic responses to Pneumocystis proteins in human immunodeficiency virus patients with and without Pneumocystis jirovecii pneumonia. J Acquir Immune Defic Syndr. 2011; 57: 190-196. [CrossRef] [Google scholar] [PubMed]
- Blount RJ, Jarlsberg LG, Daly KR, Worodria W, Davis JL, Cattamanchi A, et al. Serologic responses to recombinant Pneumocystis jirovecii major surface glycoprotein among Uganda patients with respiratory symptoms. PLoS One. 2012; 7: e51545. [CrossRef] [Google scholar] [PubMed]
- Djawe K, Daly KR, Levin L, Zar HJ, WAlzer PD. Humoral immune responses to Pneumocystis jirovecii antigens in HIV-infected and uninfected young children with Pneumocystis Pneumonia. PLoS One. 2013; 8: e82783. [CrossRef] [Google scholar] [PubMed]
- Tomás AL, Cardoso F, Esteves F, Matos O. Serological diagnosis of pneumocystosis: production of a synthetic recombinant antigen for immunodetection of Pneumocystis jirovecii. Sci Rep. 2016; 6: 36287. [CrossRef] [Google scholar] [PubMed]
- Hamada H, Kohno K, Yokoyama A, Hirasawa Y, Hiwakada K, Sakatani M, et al. KL-6 as a serologic indicator of Pneumocystis carinii pneumonia in immunocompromised hosts. Intern Med Tokyo. 1998; 37: 307-310. [CrossRef] [Google scholar] [PubMed]
- Teramoto S, Sawaki D, Okada S, Ouchi Y. Markedly increased plasma (1–3)-beta-D-glucan is a diagnostic and therapeutic indicator of Pneumocystis carinii pneumonia in a non-AIDS patient. J Med Microbiol. 2000; 49: 393-394. [CrossRef] [Google scholar] [PubMed]
- Skelly MJ, Holzman RS, Merali S. S-adenosylmethionine levels in the diagnosis of Pneumocystis carinii pneumonia in patients with HIV infection. Clin Infect Dis. 2008; 46: 467-471. [CrossRef] [Google scholar] [PubMed]
- Held J, Koch MS, Reischl U, Danner T, Serr A. Serum (1–3)-β -D-glucan measurement as an early indicator of Pneumocystis jirovecii pneumonia and evaluation of its prognostic value. Clin Microbiol Infect. 2011; 17: 595-602. [CrossRef] [Google scholar] [PubMed]
- Esteves F, Lee CH, de Sousa B, Badura R, Seringa M, Fernandes C, et al. (1–3)-beta-D-glucan in association with lactate dehydrogenase as biomarkers of Pneumocystis pneumonia (PcP) in HIV-infected patients. Eur J Clin Microbiol Infect Dis. 2014; 33: 1173-1180. [CrossRef] [Google scholar] [PubMed]
- Esteves F, Calé SS, Badura R, de Boer MG, Maltez F, Calderón EJ, et al. Diagnosis of Pneumocystis pneumonia: evaluation of four serologic biomarkers. Clin Microbiol Infect. 2015; 21: 379. e1-e10. [Google scholar]
- Karageorgopoulos DE, Qu JM, Korbila IP, Zhu YG, Vaileiou VA, Falagas ME. Accuracy of beta-d-glucan for the diagnosis of Pneumocystis jirovecii pneumonia: a meta-analysis. Clin Microbiol Infect. 2013; 19: 39-49. [CrossRef] [Google scholar] [PubMed]
- Wood BR, Komarow L, Zolopa AR, Finkelman MA, Powderly WG, Sax PE. Test performance of blood beta-glucan for Pneumocystis jirovecii pneumonia in patients with AIDS and respiratory symptoms. AIDS. 2013; 27: 967-972. [CrossRef] [Google scholar] [PubMed]
- de Boer MG, Gelinck LB, van Zelst BD, van de Sande WW, Willems LN, van Dissel JT, et al. β-D-glucan and S-adenosylmethionine serum levels for the diagnosis of Pneumocystis pneumonia in HIV-negative patients: a prospective study. J Infect. 2011; 62: 93-100. [CrossRef] [Google scholar] [PubMed]
- Nakamura H, Tateyama M, Tasato D, Haranaga S, Yara S, Higa F, et al. Clinical Utility of Serum β-D-Glucan and KL-6 Levels in Pneumocystis jirovecii Pneumonia. Intern Med. 2009; 48: 195-202. [CrossRef] [Google scholar] [PubMed]
- Damiani C, Le Gal S, Da Costa C, Virmaux M, Nevez G, Totet A. Combined quantification of pulmonary Pneumocystis jirovecii DNA and serum (1->3)-β-D-glucan for differential diagnosis of Pneumocystis pneumonia and Pneumocystis colonization. J Clin Microbiol. 2013; 51: 3380-3388. [CrossRef] [Google scholar] [PubMed]
- Matsumura Y, Ito Y, Iinuma Y, Yasuma K, Yamamoto M, Matsushima A, et al. Quantitative real-time PCR and the (1→3)-β-Dglucan assay for differentiation between Pneumocystis jirovecii pneumonia and colonization. Clin Microbiol Infect. 2012; 18: 591-597. [CrossRef] [Google scholar] [PubMed]
- Vogel M, Weissgerber P, Goeppert B, Hetzel J, Vatlach M, Claussen C, et al. Accuracy of serum LDH elevation for the diagnosis of Pneumocystis jirovecii pneumonia. Swiss Med Wkly. 2011; 141: w13184. [Google scholar]
- Skelly M, Hoffman J, Fabbri M, Holzman RS, Clarkson AB Jr, Merali S. Sadenosylmethionine concentrations in diagnosis of Pneumocystis carinii pneumonia. Lancet. 2003; 361: 1267-1268. [CrossRef] [Google scholar] [PubMed]
- Kutty G, Hernandez-Novoa B, Czapiga M, Kovaks JA. Pneumocystis encodes a functional S-adenosylmethionine synthetase gene. Eukaryot Cell. 2008; 7: 258-267. [CrossRef] [Google scholar] [PubMed]
- Onishi A, Sugiyama D, Kogata Y, Saegusa J, Sugimoto T, Kawano S, et al. Diagnostic accuracy of serum 1,3-beta-d-glucan for Pneumocystis jiroveci pneumonia, invasive candidiasis, and invasive aspergillosis: systematic review and meta-analysis. J Clin Microbiol. 2012; 50: 7-15. [CrossRef] [Google scholar] [PubMed]
- Li WJ, Guo YL, Liu TJ, Wang K, Kong JL. Diagnosis of Pneumocystis pneumonia using serum (1-3)-β-Glucan: a bivariate meta-analysis and systematis review. Thorac Dis. 2015; 7: 2214-2225. [Google scholar]
- Salerno D, Mushatt D, Myers L, Zhuang Y, de la Rua N, Calderon EJ, et al. Serum and BAL beta-D-glucan for the diagnosis of Pneumocystis pneumonia in HIV positive patients. Respir Med. 2014; 108: 1688-1695. [CrossRef] [Google scholar] [PubMed]
- Alanio A, Hauser PM, Lagrou K, Melchers WJ, Helweg-Larsen J, Matos O, et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016; 71: 2386-2396. [CrossRef] [Google scholar] [PubMed]
- Milledge J, Kakakios A, Gillis J, Fitzgerald DA. Pneumocystis carinii pneumonia as a presenting feature of X-linked hyper-IgM syndrome. J Paediatr Child Health. 2003; 39: 704-706. [CrossRef] [Google scholar] [PubMed]
- Morris A, Norris KA. Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev 2012; 25: 297-317. [CrossRef] [Google scholar] [PubMed]
- Stringer JR. Surface Antigens. In: Pneumocystis Pneumonia, 3rd edn. New York: Marcel Dekker; 2005. p. 95-126. [Google scholar]
- Dai J, Jiang M, Wang Y, Qu L, Gong R, Si J. Evaluation of a recombinant multiepitope peptide for serodiagnosis of Toxoplasma gondii infection. Clin Vaccine Immunol. 2012; 19: 338-342. [CrossRef] [Google scholar] [PubMed]
- Dai JF, Jiang M, Qu LL, Sun L, Wang YY, Gong LL, et al. Toxoplasma gondii: enzyme-linked immunosorbent assay based on a recombinant multi-epitope peptide for distinguishing recent from past infection in human sera. Exp Parasitol. 2013; 133: 95-100. [CrossRef] [Google scholar] [PubMed]
- Huang X, Xuan X, Hirata H, Yokoyama N, Xu L, Suzuki N, et al. Rapid immunochromatographic test using recombinant SAG2 for detection of antibodies against Toxoplasma gondii in cats. J Clin Microbiol. 2004; 42: 351-353. [CrossRef] [Google scholar] [PubMed]
- Esteves F, Tavares A, Costa MC, Gaspar J, Antunes F, Matos O. Genetic characterization of the UCS and Kex1 loci of Pneumocystis jirovecii. Eur J Clin Microbiol Infect Dis. 2009; 28: 175-178. [CrossRef] [Google scholar] [PubMed]
- Zheng M, Cai Y, Eddens T, Ricks DM, Kolls JK. Novel Pneumocystis antigen discovery using fungal surface proteomics. Infect Immun. 2014; 82: 2417-2423. [CrossRef] [Google scholar] [PubMed]
- Kierny MR, Cunningham TD, Kay BK. Detection of biomarkers using recombinant antibodies coupled to nanostructured platforms. Nano Rev. 2012; 3. doi: 10.3402/nano.v3i0.17240. [CrossRef] [Google scholar] [PubMed]
- Nagatani N, Tanaka R, Yuhi T, Endo T, Kerman K, Takamura Y, et al. Gold nanoparticle-based novel enhancement method for the development of highly sensitive immunochromatographic test strips. Sci Tech Adv Mater. 2006; 7: 270-275. [CrossRef] [Google scholar]
- Almeida MP, Pereira E, Baptista PV, Gomes I, Figueiredo S, Soares L, et al. Gold Nanoparticles in Analytical Chemistry. In: Comprehensive Analytical Chemistry, 1st ed. Netherlands: Elsevier; 2014. p. 529–567. [Google scholar]
- Baptista PV, Pereira E, Eaton P, Doria G, Miranda A, Gomes I, et al. Gold nanoparticles for the development of clinical diagnosis methods. Anal Bioanal Chem. 2008; 391: 943-950. [CrossRef] [Google scholar] [PubMed]
- Tomás AL, Cardoso F, Pinto M, de Almeida MP, de Sousa B, Pereira E, et al. Point-of-care innovative platform based on a serological bionanosensor for Pneumocystis pneumonia [Abstract P1219]. 28th European Congress of Clinical Microbiology and Infectious Diseases; 2018 April 21-24; Madrid, Spain. Available from: http://m.eccmidlive.org/#Abstracts/220874/ extend.